+
 +
+ +
 +
+ -
-
-
-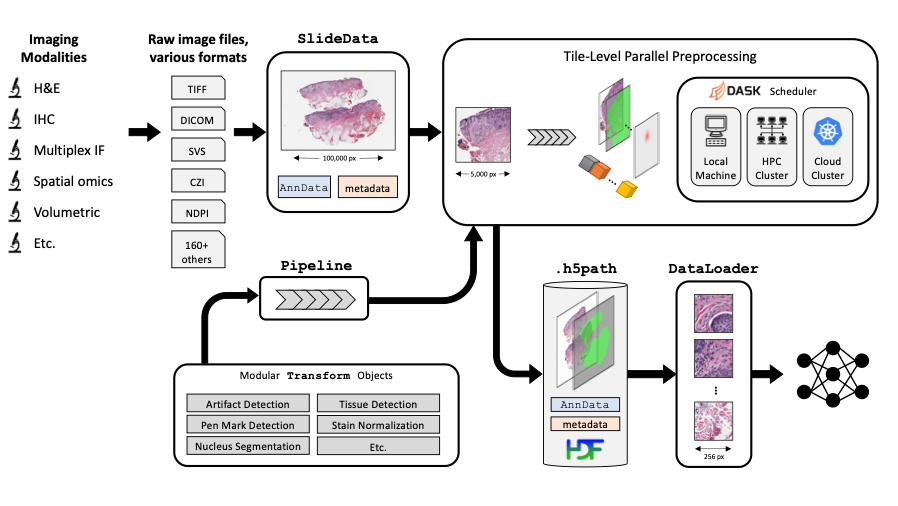 +🤖🔬 **PathML: Tools for computational pathology**
+[](https://pepy.tech/project/pathml)
[](https://pathml.readthedocs.io/en/latest/?badge=latest)
+[](https://codecov.io/gh/Dana-Farber-AIOS/pathml)
[](https://github.com/psf/black)
[](https://pypi.org/project/pathml/)
-[](https://pepy.tech/project/pathml)
-[](https://codecov.io/gh/Dana-Farber-AIOS/pathml)
+
+⭐ **PathML objective is to lower the barrier to entry to digital pathology**
+
+Imaging datasets in cancer research are growing exponentially in both quantity and information density. These massive datasets may enable derivation of insights for cancer research and clinical care, but only if researchers are equipped with the tools to leverage advanced computational analysis approaches such as machine learning and artificial intelligence. In this work, we highlight three themes to guide development of such computational tools: scalability, standardization, and ease of use. We then apply these principles to develop PathML, a general-purpose research toolkit for computational pathology. We describe the design of the PathML framework and demonstrate applications in diverse use cases.
+
+🚀 **The fastest way to get started?**
+
+ docker pull pathml/pathml && docker run -it -p 8888:8888 pathml/pathml
| Branch | Test status |
| ------ | ------------- |
| master |  |
| dev |  |
-An open-source toolkit for computational pathology and machine learning.
+
+🤖🔬 **PathML: Tools for computational pathology**
+[](https://pepy.tech/project/pathml)
[](https://pathml.readthedocs.io/en/latest/?badge=latest)
+[](https://codecov.io/gh/Dana-Farber-AIOS/pathml)
[](https://github.com/psf/black)
[](https://pypi.org/project/pathml/)
-[](https://pepy.tech/project/pathml)
-[](https://codecov.io/gh/Dana-Farber-AIOS/pathml)
+
+⭐ **PathML objective is to lower the barrier to entry to digital pathology**
+
+Imaging datasets in cancer research are growing exponentially in both quantity and information density. These massive datasets may enable derivation of insights for cancer research and clinical care, but only if researchers are equipped with the tools to leverage advanced computational analysis approaches such as machine learning and artificial intelligence. In this work, we highlight three themes to guide development of such computational tools: scalability, standardization, and ease of use. We then apply these principles to develop PathML, a general-purpose research toolkit for computational pathology. We describe the design of the PathML framework and demonstrate applications in diverse use cases.
+
+🚀 **The fastest way to get started?**
+
+ docker pull pathml/pathml && docker run -it -p 8888:8888 pathml/pathml
| Branch | Test status |
| ------ | ------------- |
| master |  |
| dev |  |
-An open-source toolkit for computational pathology and machine learning.
+ +
+
+
+ **View [documentation](https://pathml.readthedocs.io/en/latest/)**
@@ -125,6 +133,23 @@ Note that these instructions assume that there are no other processes using port
Please refer to the `Docker run` [documentation](https://docs.docker.com/engine/reference/run/) for further instructions
on accessing the container, e.g. for mounting volumes to access files on a local machine from within the container.
+## Option 4: Google Colab
+
+To get PathML running in a Colab environment:
+
+````
+!pip install openslide-python
+!apt-get install openslide-tools
+!apt-get install openjdk-8-jdk-headless -qq > /dev/null
+os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
+!update-alternatives --set java /usr/lib/jvm/java-8-openjdk-amd64/jre/bin/java
+!java -version
+!pip install pathml
+````
+
+*Thanks to all of our open-source collaborators for helping maintain these installation instructions!*
+*Please open an issue for any bugs or other problems during installation process.*
+
## CUDA
To use GPU acceleration for model training or other tasks, you must install CUDA.
@@ -191,12 +216,36 @@ See [contributing](https://github.com/Dana-Farber-AIOS/pathml/blob/master/CONTRI
# Citing
-If you use `PathML` in your work, please cite our paper:
+If you use `PathML` please cite:
+
+- [**J. Rosenthal et al., "Building tools for machine learning and artificial intelligence in cancer research: best practices and a case study with the PathML toolkit for computational pathology." Molecular Cancer Research, 2022.**](https://doi.org/10.1158/1541-7786.MCR-21-0665)
+
+So far, PathML was used in the following manuscripts:
+
+- [J. Linares et al. **Molecular Cell** 2021](https://www.cell.com/molecular-cell/fulltext/S1097-2765(21)00729-2)
+- [A. Shmatko et al. **Nature Cancer** 2022](https://www.nature.com/articles/s43018-022-00436-4)
+- [J. Pocock et al. **Nature Communications Medicine** 2022](https://www.nature.com/articles/s43856-022-00186-5)
+- [S. Orsulic et al. **Frontiers in Oncology** 2022](https://www.frontiersin.org/articles/10.3389/fonc.2022.924945/full)
+- [D. Brundage et al. **arXiv** 2022](https://arxiv.org/abs/2203.13888)
+- [A. Marcolini et al. **SoftwareX** 2022](https://www.sciencedirect.com/science/article/pii/S2352711022001558)
+- [M. Rahman et al. **Bioengineering** 2022](https://www.mdpi.com/2306-5354/9/8/335)
+- [C. Lama et al. **bioRxiv** 2022](https://www.biorxiv.org/content/10.1101/2022.09.28.509751v1.full)
+- the list continues [**here 🔗 for 2023 and onwards**](https://scholar.google.com/scholar?oi=bibs&hl=en&cites=1157052756975292108)
+
+# Users
+
+
**View [documentation](https://pathml.readthedocs.io/en/latest/)**
@@ -125,6 +133,23 @@ Note that these instructions assume that there are no other processes using port
Please refer to the `Docker run` [documentation](https://docs.docker.com/engine/reference/run/) for further instructions
on accessing the container, e.g. for mounting volumes to access files on a local machine from within the container.
+## Option 4: Google Colab
+
+To get PathML running in a Colab environment:
+
+````
+!pip install openslide-python
+!apt-get install openslide-tools
+!apt-get install openjdk-8-jdk-headless -qq > /dev/null
+os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
+!update-alternatives --set java /usr/lib/jvm/java-8-openjdk-amd64/jre/bin/java
+!java -version
+!pip install pathml
+````
+
+*Thanks to all of our open-source collaborators for helping maintain these installation instructions!*
+*Please open an issue for any bugs or other problems during installation process.*
+
## CUDA
To use GPU acceleration for model training or other tasks, you must install CUDA.
@@ -191,12 +216,36 @@ See [contributing](https://github.com/Dana-Farber-AIOS/pathml/blob/master/CONTRI
# Citing
-If you use `PathML` in your work, please cite our paper:
+If you use `PathML` please cite:
+
+- [**J. Rosenthal et al., "Building tools for machine learning and artificial intelligence in cancer research: best practices and a case study with the PathML toolkit for computational pathology." Molecular Cancer Research, 2022.**](https://doi.org/10.1158/1541-7786.MCR-21-0665)
+
+So far, PathML was used in the following manuscripts:
+
+- [J. Linares et al. **Molecular Cell** 2021](https://www.cell.com/molecular-cell/fulltext/S1097-2765(21)00729-2)
+- [A. Shmatko et al. **Nature Cancer** 2022](https://www.nature.com/articles/s43018-022-00436-4)
+- [J. Pocock et al. **Nature Communications Medicine** 2022](https://www.nature.com/articles/s43856-022-00186-5)
+- [S. Orsulic et al. **Frontiers in Oncology** 2022](https://www.frontiersin.org/articles/10.3389/fonc.2022.924945/full)
+- [D. Brundage et al. **arXiv** 2022](https://arxiv.org/abs/2203.13888)
+- [A. Marcolini et al. **SoftwareX** 2022](https://www.sciencedirect.com/science/article/pii/S2352711022001558)
+- [M. Rahman et al. **Bioengineering** 2022](https://www.mdpi.com/2306-5354/9/8/335)
+- [C. Lama et al. **bioRxiv** 2022](https://www.biorxiv.org/content/10.1101/2022.09.28.509751v1.full)
+- the list continues [**here 🔗 for 2023 and onwards**](https://scholar.google.com/scholar?oi=bibs&hl=en&cites=1157052756975292108)
+
+# Users
+
+| This is where in the world our most enthusiastic supporters are located:
+ +  +
+ |
+and this is where they work:
+ +  +
+ |
+
 diff --git a/docs/readthedocs-requirements.txt b/docs/readthedocs-requirements.txt
index 831e134d..95b9a3cc 100644
--- a/docs/readthedocs-requirements.txt
+++ b/docs/readthedocs-requirements.txt
@@ -1,7 +1,8 @@
-sphinx==4.3.2
+sphinx==7.1.2
nbsphinx==0.8.8
nbsphinx-link==1.3.0
-sphinx-rtd-theme==1.0.0
-sphinx-autoapi==1.8.4
-ipython==7.31.1
+sphinx-rtd-theme==1.3.0
+sphinx-autoapi==3.0.0
+ipython==8.10.0
sphinx-copybutton==0.4.0
+
diff --git a/environment.yml b/environment.yml
index 22754c02..0c64028f 100644
--- a/environment.yml
+++ b/environment.yml
@@ -6,12 +6,10 @@ channels:
dependencies:
- pip==21.3.1
- - python==3.8
- numpy==1.19.5
- scipy==1.7.3
- scikit-image==0.18.3
- matplotlib==3.5.1
- - python-spams==2.6.1
- openjdk==8.0.152
- pytorch==1.10.1
- h5py==3.1.0
@@ -26,7 +24,7 @@ dependencies:
- protobuf==3.20.1
- deepcell==0.11.0
- opencv-contrib-python==4.5.3.56
- - openslide-python==1.1.2
+ - openslide-python==1.3.1 # upate to openslide 4.0.0
- scanpy==1.8.2
- anndata==0.7.8
- tqdm==4.62.3
diff --git a/pathml/_version.py b/pathml/_version.py
index b91684f4..2704c284 100644
--- a/pathml/_version.py
+++ b/pathml/_version.py
@@ -3,4 +3,4 @@
License: GNU GPL 2.0
"""
-__version__ = "2.1.0"
+__version__ = "2.1.1"
diff --git a/docs/readthedocs-requirements.txt b/docs/readthedocs-requirements.txt
index 831e134d..95b9a3cc 100644
--- a/docs/readthedocs-requirements.txt
+++ b/docs/readthedocs-requirements.txt
@@ -1,7 +1,8 @@
-sphinx==4.3.2
+sphinx==7.1.2
nbsphinx==0.8.8
nbsphinx-link==1.3.0
-sphinx-rtd-theme==1.0.0
-sphinx-autoapi==1.8.4
-ipython==7.31.1
+sphinx-rtd-theme==1.3.0
+sphinx-autoapi==3.0.0
+ipython==8.10.0
sphinx-copybutton==0.4.0
+
diff --git a/environment.yml b/environment.yml
index 22754c02..0c64028f 100644
--- a/environment.yml
+++ b/environment.yml
@@ -6,12 +6,10 @@ channels:
dependencies:
- pip==21.3.1
- - python==3.8
- numpy==1.19.5
- scipy==1.7.3
- scikit-image==0.18.3
- matplotlib==3.5.1
- - python-spams==2.6.1
- openjdk==8.0.152
- pytorch==1.10.1
- h5py==3.1.0
@@ -26,7 +24,7 @@ dependencies:
- protobuf==3.20.1
- deepcell==0.11.0
- opencv-contrib-python==4.5.3.56
- - openslide-python==1.1.2
+ - openslide-python==1.3.1 # upate to openslide 4.0.0
- scanpy==1.8.2
- anndata==0.7.8
- tqdm==4.62.3
diff --git a/pathml/_version.py b/pathml/_version.py
index b91684f4..2704c284 100644
--- a/pathml/_version.py
+++ b/pathml/_version.py
@@ -3,4 +3,4 @@
License: GNU GPL 2.0
"""
-__version__ = "2.1.0"
+__version__ = "2.1.1"