Via BLAST Search panel users can perform BLAST search nucleotide/amino acid sequences over BLAST databases and view the corresponding results.
BLAST databases should be previously uploaded into NGB (this could be as downloaded NCBI databases or custom ones).
The BLAST panel contains two sub-tabs:
- Search (opened by default) - to display and specify search settings, and also start a new search
- History - to display the history of searches
To start the search, user should specify:
- The BLAST Tool that will be used for the search. Only one tool can be selected:

blastn(default) - for the search over nucleotide databases using a nucleotide queryblastp- for the search over protein databases using an amino acid queryblastx- for the search over protein databases using a translated nucleotide querytblastn- for the search over translated nucleotide databases using an amino acid querytblastx- for the search over translated nucleotide databases using a translated nucleotide query
- Query sequence, e.g.:

- The search Task title (optionally). In this field, user may specify the title for the current search operation - to easier find it later, e.g.:

- Search "set". Here user should specify where the search should be performed:
- Database - dropdown list to select the BLAST database (one from the list of all uploaded databases). The list of databases corresponds one of the types - protein or nucleotide. This type is defined by the selected tool type at the tool selector: for
blastn,tblastn,tblastx- nucleotide databases are being displayed, forblastp,blastx- protein databases are being displayed - Organism (optionally) - dropdown list to select/specify the species for which the search should be performed. Multi-select is supported.
- Separately, user can set the certain checkbox next to the Organism field to exclude specified organism(s) from the list for which the search will be performed
- Algorithm - dropdown list to select algorithms available for the current BLAST tool. Use the info icon next to that dropdown list to view a short help

- Database - dropdown list to select the BLAST database (one from the list of all uploaded databases). The list of databases corresponds one of the types - protein or nucleotide. This type is defined by the selected tool type at the tool selector: for
- Additional parameters - collapsible section to specify additional "technical" BLAST parameters that should be used for the search. Expanded view of that section:

Here:- Max target sequences - field to set the maximum number of aligned sequences to display in results (CLI BLAST option
max_target_seqs) - Expect threshold - field to set expect value for saving hits (CLI BLAST option
evalue) - Options (optionally) - field to specify additional BLAST options in CLI style. If user will try to specify such options in incorrect format or with incorrect names - they will be ignored during the search. To view the full list of supported options use the info icon next to that field
Note: Max target sequences and Expect threshold settings have default values. I.e. if user doesn't forcibly specify values for these parameters (e.g., even doesn't expand the "Additional parameters" section) - their default values shall be used for the search
- Max target sequences - field to set the maximum number of aligned sequences to display in results (CLI BLAST option
Once the BLAST search setup is finished - click the Search button to start:

Since the BLAST search can take a long time, the results are not displayed immediately - and for each search - a new "search task" is being created.
The list of such tasks is displayed at the History sub-tab.
This sub-tab is being opened automatically after the search starts:

Just-started search task is shown on the top of the tasks list.
This table contains all user's BLAST search requests.
Here:
- Task ID - automatic created ID of the certain search task
- Task title - title of the certain search task (if it was specified before the search)
- Current state - status of the search task:
- Searching - for task being performed at the moment. All new tasks get this status by default
- Done - for task successfully finished
- Interrupted - for task canceled during the searching
- Failure - for failed task (task finished with errors)
- Submitted at - date and time when the certain search was started
- Duration - duration of the certain search task
- block of the controls near each request:
- button to cancel search
 . This button allows to break the search and change the state of that task to "Interrupted". Note: available only for the task that is being currently performed
. This button allows to break the search and change the state of that task to "Interrupted". Note: available only for the task that is being currently performed - button to re-open the search setup
 . This button allows to open the Search setup sub-tab and set the search parameters in the same values as they were in the current request
. This button allows to open the Search setup sub-tab and set the search parameters in the same values as they were in the current request
- button to cancel search
- Clear history button
 at the top of the tab - to clear all BLAST search history
at the top of the tab - to clear all BLAST search history
Table supports sorting by any column (except Duration).
Table has the auto-refresh every 5 sec (only if the sub-tab is opened).
If the certain search is finished, its state changes to Done and task ID becomes a hyperlink. User can click such hyperlink (or just a row) to open the corresponding search results, e.g.:

Search results will be opened in the same tab (History):

This table contains:
- Back button (near the search task title) - to hide current results and return to the search history view:

- Header with the opened search task title (if it was specified before the search)
- BLAST parameters collapsible section (collapsed by default). This section contains details of the opened search:

- Block of additional buttons at the right-upper corner of the form:
- Sequences table - to show the search results summary grouped by sequences:

Sequences table contains BLAST search aggregated results grouped by their sequences.
Table contains the following columns:
- Sequence ID - IDs of the sequences in which hits were found
- Organism - organism specified in the sequence
- Tax ID - taxonomy ID specified in the sequence
- Max score - the highest alignment score from all matches of the search query to the certain sequence
- Total score - sum of alignment scores from all matches of the search query to the certain sequence
- Query cover - the percent of the query length that is included in the aligned segments
- E value - the number of alignments expected by chance with the calculated score or better. By this column the default sorting is set (ascending)
- Percent identity - the highest percent identity for a set of aligned segments to the same subject sequence
- Matches - number of matches of the search query to the certain sequence
Maximal count of rows in this table is defined by Max target sequences parameter that is specified before the search.
User have the ability to sort this table by any column and manually configure the column order.
User can click any row in the Sequence table and the form with details about all matches (alignments) of the search query to the certain sequence will be opened.
This form is being opened in the same tab (History), e.g.:


This form contains:
- Back button - to hide the Alignments info form and return to the Sequences table view:

- Sequence ID. ID is a hyperlink:
- if the search was performed on the NCBI database - this hyperlink opens a corresponding sequence page on NCBI
- if the search was performed on the "custom" database - this hyperlink opens a corresponding reference and its chromosome (to which this sequence belongs) in the "Browser" panel
- Details about all matches of the search query to the current sequence (match blocks)
Each match block contains:
- Range - positions of the current sequence where the match is defined
- "View at track" link - hyperlink to view the certain match (alignment) to the current sequence. This hyperlink is visible only for those sequences which references are in NGB databases
- Score and bit-score, Expect value (E-value)
- Count of Identities between sequences (by symbols) and its percent value
- Count of Gaps (by symbols) and its percent value
- Strands of each sequence (query and subject) - only for nucleotide sequences
- block with the conventional figure of the query string alignment to the current sequence segment:
- start and end position of the query string segment
- aligned query string segment
- start and end position of the current sequence segment
- the current sequence segment to which the query string segment was aligned
- symbols that "link" the corresponding letters in both sequences:
- straight line (
|) if letters are equal - nothing (empty) if letters are not equal (mismatch)
- minus symbol (
-) in any sequence - for gaps
- straight line (
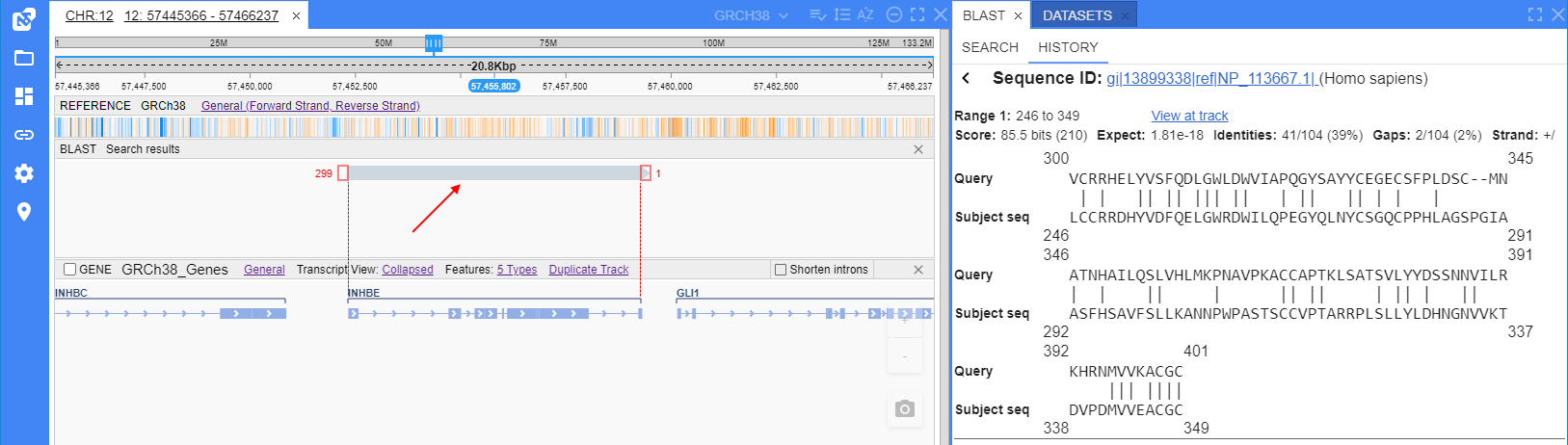
User can view any found match (alignment) at a track (graphic visualization) in the "Browser" panel. Note: it is possible only for those sequences which references are in NGB databases.
To open the visualization track, user should click the "View at track" link near the match in the "Alignments info" form:


Opened alignment is shown at the separate track.
Track is being displayed in the similar manner as a single read at the Alignment track in the "Browser":
- query matches - shown as a gray line by width of aligned matches
- query strand - shown as an arrow on the edge of the query line
- mismatches - shown as separate color rectangles with the corresponding letters
- gaps in sequence - shown as insertions (a perpendicular violet line in the gap position)
- gaps in query - shown as deletions (a black line linked two "separate" parts of the query)
- additionally, near the each end of the query line at the track, are conventionally shown counts of the query positions that were not aligned to sequence
If there are several matches (alignments) were found for the same sequence - they all will be shown at that track, e.g.:

Please note, that described behavior is valid only for searches over nucleotide sequences.
Visualization for searches over amino acid sequences does not show exact location of the alignment. In such cases, only whole gene feature is highlighted instead.
Users have the ability to start BLAST Search from the genomic feature via the "Browser" panel as well.
To prepare the search in such way, user shall just click a read (or any other feature) at a track and select any "BLAST search" option (BLASTn or BLASTp) from the context menu, e.g.:

After that, the BLAST panel will be opened. Also, some default values will be set:
- if user selected "BLASTn Search" option, at the "BLAST" panel:
blastntool type will be selected- the corresponding nucleotide sequence (according to the selected in GUI) will appear in the Query sequence field
- if user selected "BLASTp Search" option, at the "BLAST" panel:
blastptool type will be selected- the corresponding amino acids sequence (according to the selected in GUI) will appear in the Query sequence field
Example for the gene at the picture above when the "BLASTn Search" option was selected:

Note: this feature has a number of limitations. If the BLAST search can not be performed from the context menu (due to some restrictions) - the corresponding item will be disabled and a short info with a reason will be shown. E.g. if the read/feature/gene selected at the GUI for the search is too large to be inserted into the Query sequence field: