| marp | theme | style |
|---|---|---|
true |
default |
section{
justify-content: flex-start;
}
|
This is a presentation given to the engineering team at LatchBio.
- Introduction to gene editing tools
- Cas9 structure and mechanisms
- Base Editors structure and mechanisms
- State of BEs in the clinic
- Defining "Genotoxicity"
- Designing an experiment to evaluate BE behavior
Broadly, editors are molecular tools that allow us to change the DNA within cells.
They are nucleases, enzymes that cut DNA, that have been discovered and isolated from bacterial immune systems.
Editors have been engineered in a variety of ways to gain new capabilities:
- types of edits (swap letters, insertions, deletions)
- how efficient they can make edits
- how accurate they can make edits (or how little they mess up)
- how isolated their behavior is from other mechanisms in the cell
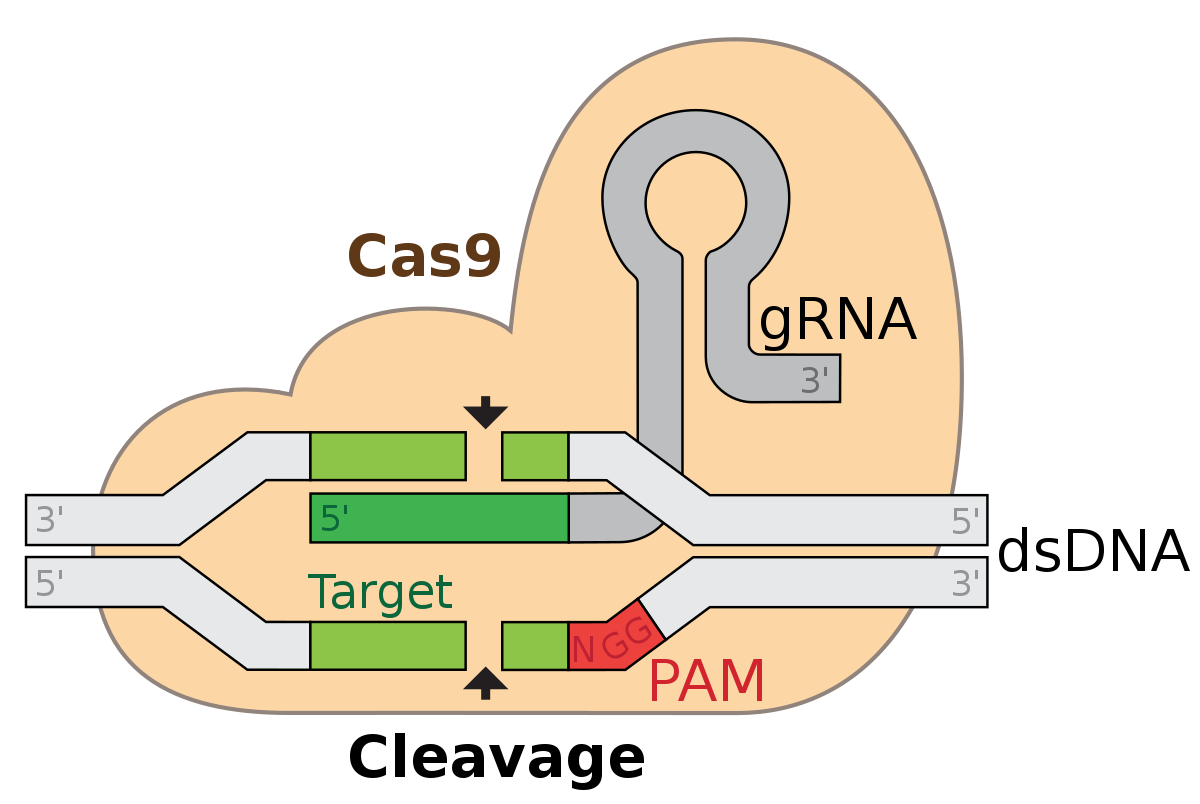
Cas9 is a nuclease isolated from bacteria (Streptococcus pyogenes).
- Paired with 20 character RNA sequence
- Allows it to bind to specific locations on the genome
- Makes double-stranded cuts of the DNA.
-
CRISPR is a location and encoding scheme for guide RNAs in the bacteria's genome.
-
[Clustered Regularly Interspaced Short Palindromic Repeats]
-
Useful guides are retrieved to protect the host against pathogens (destroy DNA)
-
Cas9 is just one of many Cas nucleases
-
We continue to discover new natural Cas proteins with unique cutting behavior and structure.
Cas9 can be used in one of two ways to alter the guide site:
- Disrupt cut site with uncontrolled insertions or deletions
- Introduce new characters to cut site
Non-homology End Joining [NHEJ]
-
Rely on existing repair mechanisms to join the disconnected ends
-
Stochastically introduces insertions or deletions that disrupt function of gene
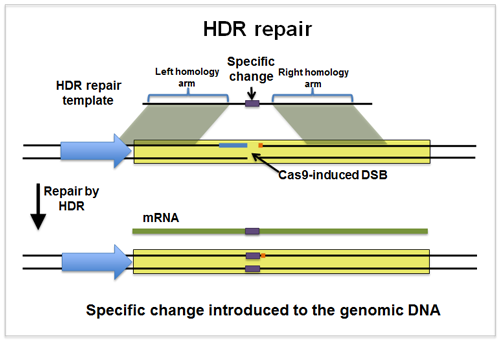
Homology Directed Repair [HDR]
A homologous sequence is a similar sequence.
Use template sequence to introduce new letters to our cut site.
-
Double-stranded breaks trigger repair pathways (p53) that can introduce large insertions, deletions, "translocations"
-
Occurs with both techniques, symptom of the DSB
-
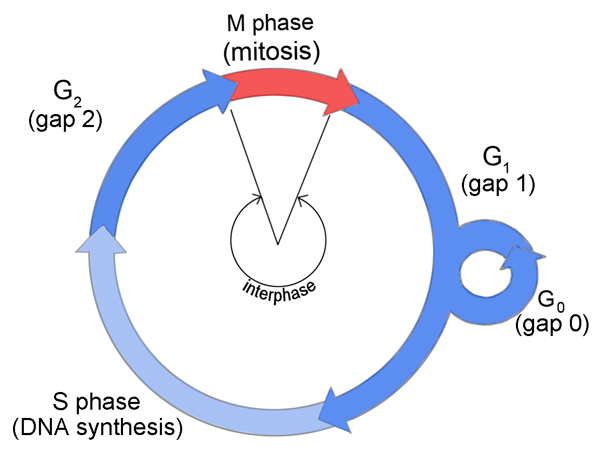
HDR can only occurs during fraction of cell cycle
Double-stranded breaks are problematic
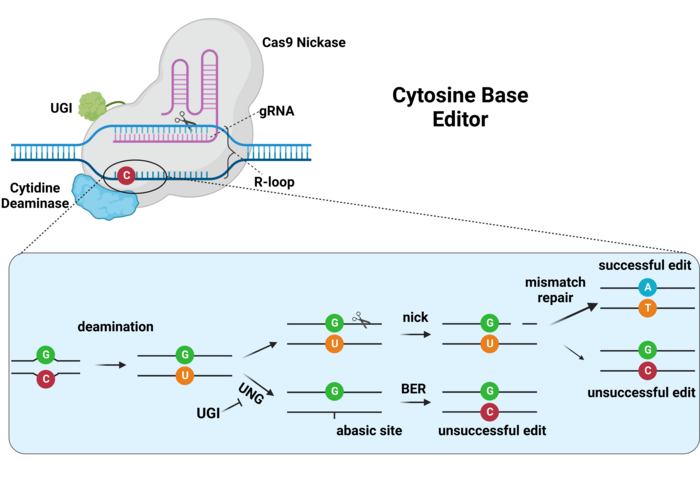
Base Editors (BE) allow conversion of single nucleotides with single stranded break.
There are two classes of BEs, defined by the nucleotide they swap
CBE (C -> T) ABE (A -> G)
They are a fusion of three proteins:
- catalytically (full or partial) inactive Cas protein
- Deaminase - chemically modifies nucleotide
- UGI - Inhibitor of a repair pathway (CBEs only)
Also called nCas.
Produce "nicks" or single-stranded breaks.
Remove amine group (NH3) turns "C" into "U".
"U" becomes "T" during DNA replication.
Inhibit natural repair of "U" back to "C".
-
No DSBs means less large insertions and deletions from repair pathways
-
Precise (single nucleotide resolution)
-
Accurate (less "off targets")
-
(Many rare diseases are caused by single nucleotide mutations)
-
Beam Therapeutics: treatment for sickle cell disease (single mutation) [Phase 1]
-
Verve Therapeutics: treatment for transthyretin amyloidosis (ATTR), a rare genetic disease [Phase 1]
-
Editas Medicine: treatment for treat Leber congenital amaurosis (LCA) [Phase1/2]
Base Editors might create unintended edits that have a toxic effect on patients.
Potential failure modes:
- Single stranded break can become a double stranded break
- "Constitutive" expression of deaminases causing havoc across genome
- Engineered components triggering bad response from cell
How do we design an experiment to measure the editing behavior of different base editors?
- Identify gene to disrupt
- Design guides to disrupt gene
- Electroporate cells with different editors (and control)
- Measure expressed protein at different time points
- Sequence the cell to observe potential misbehavior
- Expressed on the cell surface
- Easy to link gene knock out to observation (no protein)
- Measure with a flow cytometry experiment
- mRNA sequences with encoded editors are designed
- Cells are electroporated to induce uptake and expression of foreign sequence
- Y-axis: Expression of our targeted surface protein
- X-axis: The editor introduced to cell
How does a longer growing time of cell population lead to greater edit efficiency?
- Y-axis: Expression of our targeted surface protein
- X-axis: The editor introduced to cell
<style> blockquote { border-top: 0.1em dashed #555; font-size: 60%; margin-top: auto; } </style>
Review section "Optimized mRNA design improves efficiency and precision of base editing at target sites"
Explain why claims made in this paper about off target behavior, editing efficiency, recruited pathways, etc. might be inflated