SnappyData bundles Spark and supports all the Spark APIs. You can create Object based RDDs and run transformations or use the higher level APIs (like Spark ML).
All SnappyData managed tables are also accessible as DataFrame and the API extends Spark classes like SQLContext and DataFrames.
We therefore recommend that you understand the concepts in SparkSQL
and the DataFrame API. You can also store and manage arbitrary
RDDs (or even Spark DataSets) through implicit or explicit transformation to a DataFrame. While, the complete SQL support is still
evolving, the supported SQL is much richer than SparkSQL. The extension SQL supported by the SnappyStore can be referenced here.
In Spark SQL, all tables are temporary and cannot be shared across different applications. While you can manage such temporary tables, SnappyData tables are automatically registered to a built-in persistent catalog. This is similar to how Spark SQL uses the Hive catalog to natively work with Hive clusters. Data in tables is primarily managed in-memory with one or more consistent copies across machines or racks, but it can also be reliably managed on disk.
SnappySession is the main entry point for SnappyData extensions to Spark. A SnappySession extends Spark's SparkSession to work with Row and Column tables. Any DataFrame can be managed as a SnappyData table and any table can be accessed as a DataFrame. Similarly, SnappyStreamingContext is an entry point for SnappyData extensions to Spark Streaming and it extends Spark's Streaming Context.
Also SnappyData can be run in three different modes, Local Mode, Embedded Mode and SnappyData Connector mode. Before proceeding, it is important that you understand these modes. For more information, see SnappyData Spark Affinity modes.
If you are using SnappyData in LocalMode or Connector mode, it is the responsibility of the user to create a SnappySession.
###To Create a SnappySession
val spark: SparkSession = SparkSession
.builder
.appName("SparkApp")
.master("master_url")
.getOrCreate
val snappy = new SnappySession(spark.sparkContext) SparkSession spark = SparkSession
.builder()
.appName("SparkApp")
.master("master_url")
.getOrCreate();
JavaSparkContext jsc = new JavaSparkContext(spark.sparkContext());
SnappySession snappy = new SnappySession(spark.sparkContext()); from pyspark.sql.snappy import SnappySession
from pyspark import SparkContext, SparkConf
conf = SparkConf().setAppName(appName).setMaster(master)
sc = SparkContext(conf=conf)
snappy = SnappySession(sc) val spark: SparkSession = SparkSession
.builder
.appName("SparkApp")
.master("master_url")
.getOrCreate
val snsc = new SnappyStreamingContext(spark.sparkContext, Duration(1)) SparkSession spark = SparkSession
.builder()
.appName("SparkApp")
.master("master_url")
.getOrCreate();
JavaSparkContext jsc = new JavaSparkContext(spark.sparkContext());
Duration batchDuration = Milliseconds.apply(500);
JavaSnappyStreamingContext jsnsc = new JavaSnappyStreamingContext(jsc, batchDuration); from pyspark.streaming.snappy.context import SnappyStreamingContext
from pyspark import SparkContext, SparkConf
conf = SparkConf().setAppName(appName).setMaster(master)
sc = SparkContext(conf=conf)
duration = .5
snsc = SnappyStreamingContext(sc, duration)If you are in the Embedded Mode, applications typically submit Jobs to SnappyData and do not explicitly create a SnappySession or SnappyStreamingContext. These jobs are the primary mechanism to interact with SnappyData using the Spark API. A job implements either SnappySQLJob or SnappyStreamingJob (for streaming applications) trait.
The implementation of the runSnappyJob function from SnappySQLJob uses a SnappySession to interact with the SnappyData store to process and store tables. The implementation of runSnappyJob from SnappyStreamingJob uses a SnappyStreamingContext to create streams and manage the streaming context. The jobs are submitted to the lead node of SnappyData over REST API using a spark-submit like utility.
To create a job that can be submitted through the job server, the job must implement the SnappySQLJob or SnappyStreamingJob trait. Your job is displayed as:
class SnappySampleJob implements SnappySQLJob {
/** SnappyData uses this as an entry point to execute SnappyData jobs. **/
def runSnappyJob(snappy: SnappySession, jobConfig: Config): Any
/** SnappyData calls this function to validate the job input and reject invalid job requests **/
def isValidJob(snappy: SnappySession, config: Config): SnappyJobValidation
}class SnappySampleJob extends SnappySQLJob {
/** SnappyData uses this as an entry point to execute SnappyData jobs. **/
public Object runSnappyJob(SnappySession snappy, Config jobConfig) {//Implementation}
/** SnappyData calls this function to validate the job input and reject invalid job requests **/
public SnappyJobValidation isValidJob(SnappySession snappy, Config config) {//validate}
}class SnappyStreamingSampleJob implements SnappyStreamingJob {
/** SnappyData uses this as an entry point to execute SnappyData jobs. **/
def runSnappyJob(sc: SnappyStreamingContext, jobConfig: Config): Any
/** SnappyData calls this function to validate the job input and reject invalid job requests **/
def isValidJob(sc: SnappyStreamingContext, config: Config): SnappyJobValidation
}class SnappyStreamingSampleJob extends JavaSnappyStreamingJob {
/** SnappyData uses this as an entry point to execute SnappyData jobs. **/
public Object runSnappyJob(JavaSnappyStreamingContext snsc, Config jobConfig) {//implementation }
/** SnappyData calls this function to validate the job input and reject invalid job requests **/
public SnappyJobValidation isValidJob(JavaSnappyStreamingContext snc, Config jobConfig)
{//validate}
}Note: The Job traits are simply extensions of the SparkJob implemented by Spark JobServer.
-
runSnappyJobcontains the implementation of the Job. The SnappySession/SnappyStreamingContext is managed by the SnappyData Leader (which runs an instance of Spark JobServer) and is provided to the job through this method. This relieves the developer from the boiler-plate configuration management that comes with the creation of a Spark job and allows the Job Server to manage and re-use contexts. -
isValidJoballows for an initial validation of the context and any provided configuration. If the context and configuration can run the job, returningspark.jobserver.SnappyJobValidallows the job to execute, otherwise returningspark.jobserver.SnappyJobInvalid<reason>prevents the job from running and provides means to convey the reason of failure. In this case, the call immediately returns an "HTTP/1.1 400 Bad Request" status code. Validate helps you prevent running jobs that eventually fail due to a missing or wrong configuration, and saves both time and resources.
See examples for Spark and Spark streaming jobs.
SnappySQLJob trait extends the SparkJobBase trait. It provides users the singleton SnappyContext object that may be reused across jobs. SnappyContext singleton object creates one SQLContext per incoming SQL connection. Similarly, SnappyStreamingJob provides users access to SnappyStreamingContext object that can be reused across jobs.
The following command submits CreateAndLoadAirlineDataJob. This job creates DataFrames from parquet files, loads the data from DataFrame into column tables and row tables, and creates sample table on column table in its runJob method.
The program is compiled into a jar file (quickstart.jar) and submitted to jobs server as shown below.
$ bin/snappy-job.sh submit \
--lead hostNameOfLead:8090 \
--app-name airlineApp \
--class io.snappydata.examples.CreateAndLoadAirlineDataJob \
--app-jar $SNAPPY_HOME/examples/jars/quickstart.jarThe utility snappy-job.sh submits the job and returns a JSON that has a Job Id of this job.
-
--lead: Specifies the host name of the lead node along with the port on which it accepts jobs (8090) -
--app-name: Specifies the name given to the submitted application -
--class: Specifies the name of the class that contains implementation of the Spark job to be run -
--app-jar: Specifies the jar file that packages the code for Spark job
The status returned by the utility is displayed below:
{
"status": "STARTED",
"result": {
"jobId": "321e5136-4a18-4c4f-b8ab-f3c8f04f0b48",
"context": "snappyContext1452598154529305363"
}
}This Job ID can be used to query the status of the running job.
$ bin/snappy-job.sh status \
--lead hostNameOfLead:8090 \
--job-id 321e5136-4a18-4c4f-b8ab-f3c8f04f0b48
{
"duration": "17.53 secs",
"classPath": "io.snappydata.examples.CreateAndLoadAirlineDataJob",
"startTime": "2016-01-12T16:59:14.746+05:30",
"context": "snappyContext1452598154529305363",
"result": "See /home/user1/snappyhome/work/localhost-lead-1/CreateAndLoadAirlineDataJob.out",
"status": "FINISHED",
"jobId": "321e5136-4a18-4c4f-b8ab-f3c8f04f0b48"
}Once the tables are created, they can be queried by running another job. Please refer to AirlineDataJob for implementing the job.
$ bin/snappy-job.sh submit \
--lead hostNameOfLead:8090 \
--app-name airlineApp \
--class io.snappydata.examples.AirlineDataJob \
--app-jar $SNAPPY_HOME/examples/jars/quickstart.jarThe status of this job can be queried in the same manner as shown above. The result of the job returns a file path that has the query results.
Python users can submit a Python application using spark-submit in the SnappyData Connector mode. For example, run the command given below to submit a Python application:
$ bin/spark-submit \
--master local[*]
--conf snappydata.store.locators=localhost:10334 \
--conf spark.ui.port=4042
quickstart/python/AirlineDataPythonApp.pysnappydata.store.locators property denotes the locator URL of the SnappyData cluster and it is used to connect to the SnappyData cluster.
An implementation of SnappyStreamingJob can be submitted to the lead node of SnappyData cluster by specifying --stream as an option to the submit command. This option creates a new SnappyStreamingContext before the job is submitted.
Alternatively, you can specify the name of an existing/pre-created streaming context as --context <context-name> with the submit command.
For example, TwitterPopularTagsJob can be submitted as follows. This job creates stream tables on tweet streams, registers continuous queries and prints results of queries such as top 10 hash tags of last two second, top 10 hash tags until now, and top 10 popular tweets.
$ bin/snappy-job.sh submit \
--lead hostNameOfLead:8090 \
--app-name airlineApp \
--class io.snappydata.examples.TwitterPopularTagsJob \
--app-jar $SNAPPY_HOME/examples/jars/quickstart.jar \
--stream
{
"status": "STARTED",
"result": {
"jobId": "982ac142-3550-41e1-aace-6987cb39fec8",
"context": "snappyStreamingContext1463987084945028747"
}
}To start another streaming job with a new streaming context, you need to first stop the currently running streaming job, followed by its streaming context.
$ bin/snappy-job.sh stop \
--lead hostNameOfLead:8090 \
--job-id 982ac142-3550-41e1-aace-6987cb39fec8
$ bin/snappy-job.sh listcontexts \
--lead hostNameOfLead:8090
["snappyContext1452598154529305363", "snappyStreamingContext1463987084945028747", "snappyStreamingContext"]
$ bin/snappy-job.sh stopcontext snappyStreamingContext1463987084945028747 \
--lead hostNameOfLead:8090##Managing JAR Files
SnappyData provides system procedures that you can use to install and manage JAR files from a client connection. These can be used to install your custom code (for example code shared across multiple jobs) in SnappyData cluster.
Installing a JAR Use SQLJ.INSTALL_JAR procedure to install a JAR file
Syntax:
SQLJ.INSTALL_JAR(IN JAR_FILE_PATH VARCHAR(32672), IN QUALIFIED_JAR_NAME VARCHAR(32672), IN DEPLOY INTEGER)- JAR_FILE_PATH is the full path for the JAR file. This path must be accessible to the server on which the INSTALL_JAR procedure is being executed. If the JDBC client connection on which this procedure is being executed is using a locator to connect to the cluster, then actual client connection could be with any available servers. In this case, the JAR file path should be available to all servers
- QUALIFIED_JAR_NAME: The SnappyData name of the JAR file, qualified by a valid schema name.
- DEPLOY: This argument is currently ignored.
Example: Installing a JAR**
snappy> call sqlj.install_jar('/path_to_jar/procs.jar', 'APP.custom_procs', 0);Replacing a JAR Use SQLJ.REPLACE_JAR procedure to replace an installed JAR file
Syntax:
SQLJ.REPLACE_JAR(IN JAR_FILE_PATH VARCHAR(32672), IN QUALIFIED_JAR_NAME VARCHAR(32672))- JAR_FILE_PATH is full path for the JAR file. This path must be accessible to server on which the INSTALL_JAR procedure is being executed. If the JDBC client connection on which this procedure is being executed is using locator to connect to the cluster, then actual client connection could be with any available servers. In this case, the JAR file path should be available to all servers
- QUALIFIED_JAR_NAME: The SnappyData name of the JAR file, qualified by a valid schema name.
Example: Replacing a JAR
CALL sqlj.replace_jar('/path_to_jar/newprocs.jar', 'APP.custom_procs')Removing a JAR Use SQLJ.REMOVE_JAR procedure to remove a JAR file
Syntax:
SQLJ.REMOVE_JAR(IN QUALIFIED_JAR_NAME VARCHAR(32672), IN UNDEPLOY INTEGER)- QUALIFIED_JAR_NAME: The SnappyData name of the JAR file, qualified by a valid schema name.
- UNDEPLOY: This argument is currently ignored.
Example: Removing a JAR
CALL SQLJ.REMOVE_JAR('APP.custom_procs', 0)The SnappyData SQL Shell (snappy-sql) provides a simple command line interface to the SnappyData cluster. It allows you to run interactive queries on row and column stores, run administrative operations and run status commands on the cluster. Internally, it uses JDBC to interact with the cluster. You can also use tools like SquirrelSQL or DBVisualizer( JDBC to connect to the cluster) to interact with SnappyData.
// from the SnappyData base directory
$ cd quickstart/scripts
$ ../../bin/snappy-sql
Version 2.0-BETA
snappy>
//Connect to the cluster as a client
snappy> connect client 'localhost:1527'; //It connects to the locator.
//Show active connections
snappy> show connections;
//Display cluster members by querying a system table
snappy> select id, kind, status, host, port from sys.members;
//or
snappy> show members;
//Run a sql script. This particular script creates and loads a column table in the default schema
snappy> run 'create_and_load_column_table.sql';
//Run a sql script. This particular script creates and loads a row table in the default schema
snappy> run 'create_and_load_row_table.sql';
The complete list of commands available through snappy_shell can be found here
SnappyData, out-of-the-box, collocates Spark executors and the SnappyData store for efficient data intensive computations. You however may need to isolate the computational cluster for other reasons. For instance, a computationally intensive Map-reduce machine learning algorithm that needs to iterate over a cached data set repeatedly.
To support such cases it is also possible to run native Spark jobs that accesses a SnappyData cluster as a storage layer in a parallel fashion. To connect to the SnappyData store the spark.snappydata.store.locators property should be provided while starting the Spark-shell.
To run all SnappyData functionalities you need to create a SnappySession.
// from the SnappyData base directory
# Start the Spark shell in local mode. Pass SnappyData's locators host:port as a conf parameter.
$ bin/spark-shell --master local[*] --conf spark.snappydata.store.locators=locatorhost:port --conf spark.ui.port=4041
scala>
#Try few commands on the spark-shell. Following command shows the tables created using the snappy-sql
scala> val snappy = new org.apache.spark.sql.SnappySession(spark.sparkContext)
scala> val airlineDF = snappy.table("airline").show
scala> val resultset = snappy.sql("select * from airline")Any Spark application can also use the SnappyData as store and Spark as a computational engine by providing an extra spark.snappydata.store.locators property in the conf.
# Start the Spark standalone cluster from SnappyData base directory
$ sbin/start-all.sh
# Submit AirlineDataSparkApp to Spark Cluster with snappydata's locator host port.
$ bin/spark-submit --class io.snappydata.examples.AirlineDataSparkApp --master spark://masterhost:7077 --conf spark.snappydata.store.locators=locatorhost:port --conf spark.ui.port=4041 $SNAPPY_HOME/examples/jars/quickstart.jar
# The results can be seen on the command line.SnappyData is shipped with few JDBC drivers. The connection URL typically points to one of the locators. In the background, the driver acquires the endpoints for all the servers in the cluster along with load information, and automatically connects clients to one of the data servers directly. The driver provides HA by automatically adjusting underlying physical connections in case the servers fail.
// 1527 is the default port a Locator or Server uses to listen for thin client connections
Connection c = DriverManager.getConnection ("jdbc:snappydata://locatorHostName:1527/");
// While, clients typically just point to a locator, you could also directly point the
// connection at a server endpointNote: If the tool does not automatically select a driver class, you may have the option of selecting a class from within the JAR file. In this case, select the io.snappydata.jdbc.ClientDriver class.
SnappyData provides support for Apache Thrift protocol which enables users to access the cluster from other languages that are not supported directly by SnappyData. Thrift allows efficient and reliable communication across programming languages like Java, Python, PHP, Ruby, Elixir, Perl and other languages. For more information on Thrift, refer to the Apache Thrift documentation.
The JDBC driver for SnappyData that uses the jdbc:snappydata:// URL schema, now uses Thrift for underlying protocol. The older URL scheme for RowStore jdbc:gemfirexd:// continues to use the deprecated DRDA protocol.
Likewise, locators and servers in SnappyData now default to starting up thrift servers and when started in RowStore mode (snappy-start-all.sh rowstore) the DRDA servers are started as before.
To explicitly start a DRDA server in SnappyData, you can use the -drda-server-address and -drda-server-port options for the bind address and port respectively. Likewise, to explicitly start a Thrift server in RowStore mode, you can use the -thrift-server-address and -thrift-server-port options.
Refer to the following documents for information on support provided by SnappyData:
-
About SnappyData Thrift: Contains detailed information about the feature and it's capabilities.
-
The Thrift Interface Definition Language (IDL): This is a Thrift interface definition file for the SnappyData service.
-
Example: Example of the Thrift definitions using the SnappyData Thrift IDL.
Other than create and drop table, rest are all based on the Spark SQL Data Source APIs.
val props = Map("BUCKETS" -> "2")// Number of partitions to use in the SnappyStore
case class Data(COL1: Int, COL2: Int, COL3: Int)
val data = Seq(Seq(1, 2, 3), Seq(7, 8, 9), Seq(9, 2, 3), Seq(4, 2, 3), Seq(5, 6, 7))
val rdd = spark.sparkContext.parallelize(data, data.length).map(s => new Data(s(0), s(1), s(2)))
val df = snappy.createDataFrame(rdd)
// create a column table
snappy.dropTable("COLUMN_TABLE", ifExists = true)
// "column" is the table format (that is row or column)
// dataDF.schema provides the schema for table
snappy.createTable("COLUMN_TABLE", "column", df.schema, props)
// append dataDF into the table
df.write.insertInto("COLUMN_TABLE")
val results = snappy.sql("SELECT * FROM COLUMN_TABLE")
println("contents of column table are:")
results.foreach(r => println(r)) Map<String, String> props1 = new HashMap<>();
props1.put("buckets", "11");
JavaRDD<Row> jrdd = jsc.parallelize(Arrays.asList(
RowFactory.create(1, 2, 3),
RowFactory.create(7, 8, 9),
RowFactory.create(9, 2, 3),
RowFactory.create(4, 2, 3),
RowFactory.create(5, 6, 7)
));
StructType schema = new StructType(new StructField[]{
new StructField("col1", DataTypes.IntegerType, false, Metadata.empty()),
new StructField("col2", DataTypes.IntegerType, false, Metadata.empty()),
new StructField("col3", DataTypes.IntegerType, false, Metadata.empty()),
});
Dataset<Row> df = snappy.createDataFrame(jrdd, schema);
// create a column table
snappy.dropTable("COLUMN_TABLE", true);
// "column" is the table format (that is row or column)
// dataDF.schema provides the schema for table
snappy.createTable("COLUMN_TABLE", "column", df.schema(), props1, false);
// append dataDF into the table
df.write().insertInto("COLUMN_TABLE");
Dataset<Row> results = snappy.sql("SELECT * FROM COLUMN_TABLE");
System.out.println("contents of column table are:");
for (Row r : results.select("col1", "col2", "col3"). collectAsList()) {
System.out.println(r);
}from pyspark.sql.types import *
data = [(1,2,3),(7,8,9),(9,2,3),(4,2,3),(5,6,7)]
rdd = sc.parallelize(data)
schema=StructType([StructField("col1", IntegerType()),
StructField("col2", IntegerType()),
StructField("col3", IntegerType())])
dataDF = snappy.createDataFrame(rdd, schema)
# create a column table
snappy.dropTable("COLUMN_TABLE", True)
#"column" is the table format (that is row or column)
#dataDF.schema provides the schema for table
snappy.createTable("COLUMN_TABLE", "column", dataDF.schema, True, buckets="11")
#append dataDF into the table
dataDF.write.insertInto("COLUMN_TABLE")
results1 = snappy.sql("SELECT * FROM COLUMN_TABLE")
print("contents of column table are:")
results1.select("col1", "col2", "col3"). show()The optional BUCKETS attribute specifies the number of partitions or buckets to use. In SnappyStore, when data migrates between nodes (say if the cluster is expanded) a bucket is the smallest unit that can be moved around.
For more details about the properties ('props1' map in above example) and createTable API refer to documentation for row and column tables.
// create a row format table called ROW_TABLE
snappy.dropTable("ROW_TABLE", ifExists = true)
// "row" is the table format
// dataDF.schema provides the schema for table
val props2 = Map.empty[String, String]
snappy.createTable("ROW_TABLE", "row", dataDF.schema, props2)
// append dataDF into the data
dataDF.write.insertInto("ROW_TABLE")
val results2 = snappy.sql("select * from ROW_TABLE")
println("contents of row table are:")
results2.foreach(println)
// row tables can be mutated
// for example update "ROW_TABLE" and set col3 to 99 where
// criteria "col3 = 3" is true using update API
snappy.update("ROW_TABLE", "COL3 = 3", org.apache.spark.sql.Row(99), "COL3" )
val results3 = snappy.sql("SELECT * FROM ROW_TABLE")
println("contents of row table are after setting col3 = 99 are:")
results3.foreach(println)
// update rows using sql update statement
snappy.sql("UPDATE ROW_TABLE SET COL1 = 100 WHERE COL3 = 99")
val results4 = snappy.sql("SELECT * FROM ROW_TABLE")
println("contents of row table are after setting col1 = 100 are:")
results4.foreach(println)SnappyData extends Spark streaming so stream definitions can be declaratively written using SQL and these streams can be analyzed using static and dynamic SQL.
Below example shows how to use the SnappyStreamingContext to apply a schema to existing DStream and then query the SchemaDStream with simple SQL. It also shows the ability of the SnappyStreamingContext to deal with SQL queries.
import org.apache.spark.sql._
import org.apache.spark.streaming._
import scala.collection.mutable
import org.apache.spark.rdd._
import org.apache.spark.sql.types._
import scala.collection.immutable.Map
val snsc = new SnappyStreamingContext(spark.sparkContext, Duration(1))
val schema = StructType(List(StructField("id", IntegerType) ,StructField("text", StringType)))
case class ShowCaseSchemaStream (loc:Int, text:String)
snsc.snappyContext.dropTable("streamingExample", ifExists = true)
snsc.snappyContext.createTable("streamingExample", "column", schema, Map.empty[String, String] , false)
def rddList(start:Int, end:Int) = sc.parallelize(start to end).map(i => ShowCaseSchemaStream( i, s"Text$i"))
val dstream = snsc.queueStream[ShowCaseSchemaStream](
mutable.Queue(rddList(1, 10), rddList(10, 20), rddList(20, 30)))
val schemaDStream = snsc.createSchemaDStream(dstream )
schemaDStream.foreachDataFrame(df => {
df.write.format("column").
mode(SaveMode.Append).
options(Map.empty[String, String]).
saveAsTable("streamingExample") })
snsc.start()
snsc.sql("select count(*) from streamingExample").show StructType schema = new StructType(new StructField[]{
new StructField("id", DataTypes.IntegerType, false, Metadata.empty()),
new StructField("text", DataTypes.StringType, false, Metadata.empty())
});
Map<String, String> props = Collections.emptyMap();
jsnsc.snappySession().dropTable("streamingExample", true);
jsnsc.snappySession().createTable("streamingExample", "column", schema, props, false);
Queue<JavaRDD<ShowCaseSchemaStream>> rddQueue = new LinkedList<>();// Define a JavaBean named ShowCaseSchemaStream
rddQueue.add(rddList(jsc, 1, 10));
rddQueue.add(rddList(jsc, 10, 20));
rddQueue.add(rddList(jsc, 20, 30));
//rddList methods is defined as
/* private static JavaRDD<ShowCaseSchemaStream> rddList(JavaSparkContext jsc, int start, int end){
List<ShowCaseSchemaStream> objs = new ArrayList<>();
for(int i= start; i<=end; i++){
objs.add(new ShowCaseSchemaStream(i, String.format("Text %d",i)));
}
return jsc.parallelize(objs);
}*/
JavaDStream<ShowCaseSchemaStream> dStream = jsnsc.queueStream(rddQueue);
SchemaDStream schemaDStream = jsnsc.createSchemaDStream(dStream, ShowCaseSchemaStream.class);
schemaDStream.foreachDataFrame(new VoidFunction<Dataset<Row>>() {
@Override
public void call(Dataset<Row> df) {
df.write().insertInto("streamingExample");
}
});
jsnsc.start();
jsnsc.sql("select count(*) from streamingExample").show();from pyspark.streaming.snappy.context import SnappyStreamingContext
from pyspark.sql.types import *
def rddList(start, end):
return sc.parallelize(range(start, end)).map(lambda i : ( i, "Text" + str(i)))
def saveFunction(df):
df.write.format("column").mode("append").saveAsTable("streamingExample")
schema=StructType([StructField("loc", IntegerType()),
StructField("text", StringType())])
snsc = SnappyStreamingContext(sc, 1)
dstream = snsc.queueStream([rddList(1,10) , rddList(10,20), rddList(20,30)])
snsc._snappycontext.dropTable("streamingExample" , True)
snsc._snappycontext.createTable("streamingExample", "column", schema)
schemadstream = snsc.createSchemaDStream(dstream, schema)
schemadstream.foreachDataFrame(lambda df: saveFunction(df))
snsc.start()
time.sleep(1)
snsc.sql("select count(*) from streamingExample").show()
Column tables organize and manage data in memory in compressed columnar form such that, modern day CPUs can traverse and run computations like a sum or an average really fast (as the values are available in contiguous memory). Column table follows the Spark DataSource access model.
Row tables, unlike column tables, are laid out one row at a time in contiguous memory. Rows are typically accessed using keys and its location is determined by a hash function and hence is fast for point lookups or updates.
Create table DDL for Row and Column tables allows tables to be partitioned on primary keys, custom partitioned, replicated, carry indexes in memory, persist to disk, overflow to disk, be replicated for HA, etc.
CREATE TABLE [IF NOT EXISTS] table_name
(
COLUMN_DEFININTION
)
USING row | column
OPTIONS (
COLOCATE_WITH 'table_name', // Default none
PARTITION_BY 'PRIMARY KEY | column name', // If not specified it will be a replicated table.
BUCKETS 'NumPartitions', // Default 113
REDUNDANCY '1' ,
EVICTION_BY ‘LRUMEMSIZE 200 | LRUCOUNT 200 | LRUHEAPPERCENT,
PERSISTENT ‘ASYNCHRONOUS | SYNCHRONOUS’,
DISKSTORE 'DISKSTORE_NAME', //empty string maps to default diskstore
EXPIRE ‘TIMETOLIVE in seconds',
COLUMN_BATCH_SIZE '32000000',
COLUMN_MAX_DELTA_ROWS '10000',
)
[AS select_statement];
DROP TABLE [IF EXISTS] table_nameRefer to the How-Tos section for more information on partitioning and collocating data.
For row format tables column definition can take underlying GemFire XD syntax to create a table. For example, note the PRIMARY KEY clause below.
snappy.sql("CREATE TABLE tableName (Col1 INT NOT NULL PRIMARY KEY, Col2 INT, Col3 INT)
USING row options(BUCKETS '5')" )For column table it is restricted to Spark syntax for column definition
snappy.sql("CREATE TABLE tableName (Col1 INT ,Col2 INT, Col3 INT) USING column options(BUCKETS '5')" )You can also define complex types (Map, Array and StructType) as columns for column tables.
snappy.sql("CREATE TABLE tableName (
col1 INT ,
col2 Array<Decimal>,
col3 Map<Timestamp, Struct<x: Int, y: String, z: Decimal(10,5)>>,
col6 Struct<a: Int, b: String, c: Decimal(10,5)>
) USING column options(BUCKETS '5')" )To access the complex data from JDBC you can see JDBCWithComplexTypes for examples.
Note : Clauses like PRIMARY KEY, NOT NULL etc. are not supported for column definition.
Get a reference to SnappySession:
val snappy: SnappySession = new SnappySession(spark.sparkContext)
Create a SnappyStore table using Spark APIs
val props = Map('BUCKETS','5') //This map should contain required DDL extensions, see next section
case class Data(col1: Int, col2: Int, col3: Int)
val data = Seq(Seq(1, 2, 3), Seq(7, 8, 9), Seq(9, 2, 3), Seq(4, 2, 3), Seq(5, 6, 7))
val rdd = sparkContext.parallelize(data, data.length).map(s => new Data(s(0), s(1), s(2)))
val dataDF = snappy.createDataFrame(rdd)
snappy.createTable("column_table", "column", dataDF.schema, props)
//or create a row format table
snappy.createTable("row_table", "row", dataDF.schema, props)
Drop a SnappyStore table using Spark APIs:
snappy.dropTable(tableName, ifExists = true)
The below mentioned DDL extensions are required to configure a table based on user requirements. One can specify one or more options to create the kind of table one wants. If no option is specified, default values are attached. See next section for various restrictions.
-
COLOCATE_WITH: The COLOCATE_WITH clause specifies a partitioned table with which the new partitioned table must be colocated. The referenced table must already exist.
-
PARTITION_BY: Use the PARTITION_BY {COLUMN} clause to provide a set of column names that determines the partitioning. As a shortcut you can use PARTITION BY PRIMARY KEY to refer to the primary key columns defined for the table. If not specified, it is a replicated table.
-
BUCKETS: The optional BUCKETS attribute specifies the fixed number of "buckets," the smallest unit of data containment for the table that can be moved around. Data in a single bucket resides and moves together. If not specified, the number of buckets defaults to 113.
-
REDUNDANCY: Use the REDUNDANCY clause to specify the number of redundant copies that should be maintained for each partition, to ensure that the partitioned table is highly available even if members fail.
-
EVICTION_BY: Use the EVICTION_BY clause to evict rows automatically from the in-memory table based on different criteria. You can use this clause to create an overflow table where evicted rows are written to a local SnappyStore disk store
-
PERSISTENT: When you specify the PERSISTENT keyword, GemFire XD persists the in-memory table data to a local GemFire XD disk store configuration. SnappyStore automatically restores the persisted table data to memory when you restart the member.
-
DISKSTORE: The disk directory where you want to persist the table data. For more information, refer to this document.
-
EXPIRE: You can use the EXPIRE clause with tables to control the SnappyStore memory usage. It expires the rows after configured TTL.
-
COLUMN_BATCH_SIZE: The default size of blocks to use for storage in the SnappyData column store. When inserting data into the column storage this is the unit (in bytes) that is used to split the data into chunks for efficient storage and retrieval. The default value is 25165824 (24M)
-
COLUMN_MAX_DELTA_ROWS: The maximum number of rows that can be in the delta buffer of a column table for each bucket, before it is flushed into the column store. Although the size of column batches is limited by
COLUMN_BATCH_SIZE(and thus limits size of row buffer for each bucket as well), this property allows a lower limit on the number of rows for better scan performance. The default value is 10000.
Note: The following corresponding SQLConf properties forCOLUMN_BATCH_SIZEandCOLUMN_MAX_DELTA_ROWSare set if the table creation is done in that session (and the properties have not been explicitly specified in the DDL):
*snappydata.column.batchSize- explicit batch size for this session for bulk insert operations. If a table is created in the session without any explicitCOLUMN_BATCH_SIZEspecification, then this is inherited for that table property.
*snappydata.column.maxDeltaRows- maximum limit on rows in the delta buffer for each bucket of column table in this session. If a table is created in the session without any explicitCOLUMN_MAX_DELTA_ROWSspecification, then this is inherited for that table property.
Refer to the SQL Reference Guide for information on the extensions.
-
Column tables cannot specify any primary key, unique key constraints
-
Index on column table is not supported
-
Option EXPIRE is not applicable for column tables
-
Option EVICTION_BY with value LRUCOUNT is not applicable for column tables
INSERT OVERWRITE TABLE tablename1 select_statement1 FROM from_statement;
INSERT INTO TABLE tablename1 select_statement1 FROM from_statement;
INSERT INTO TABLE tablename1 VALUES (value1, value2 ..) ;
UPDATE tablename SET column = value [, column = value ...] [WHERE expression]
PUT INTO tableName (column, ...) VALUES (value, ...)
DELETE FROM tablename1 [WHERE expression]
TRUNCATE TABLE tablename1;
Several APIs have been added in SnappySession to manipulate data stored in row and column format. Apart from SQL these APIs can be used to manipulate tables.
// Applicable for both row and column tables
def insert(tableName: String, rows: Row*): Int .
// Only for row tables
def put(tableName: String, rows: Row*): Int
def update(tableName: String, filterExpr: String, newColumnValues: Row,
updateColumns: String*): Int
def delete(tableName: String, filterExpr: String): Int
Usage SnappySession.insert(): Insert one or more [[org.apache.spark.sql.Row]] into an existing table
val data = Seq(Seq(1, 2, 3), Seq(7, 8, 9), Seq(9, 2, 3), Seq(4, 2, 3),
Seq(5, 6, 7), Seq(1,100,200))
data.map { r =>
snappy.insert("tableName", Row.fromSeq(r))
}
Usage SnappySession.put(): Upsert one or more [[org.apache.spark.sql.Row]] into an existing table
val data = Seq(Seq(1, 2, 3), Seq(7, 8, 9), Seq(9, 2, 3), Seq(4, 2, 3),
Seq(5, 6, 7), Seq(1,100,200))
data.map { r =>
snappy.put(tableName, Row.fromSeq(r))
}
Usage SnappySession.update(): Update all rows in table that match passed filter expression
snappy.update(tableName, "ITEMREF = 3" , Row(99) , "ITEMREF" )
Usage SnappySession.delete(): Delete all rows in table that match passed filter expression
snappy.delete(tableName, "ITEMREF = 3")
SnappyData supports CHAR and VARCHAR datatypes in addition to Spark's String datatype. For performance reasons, it is recommended that you use either CHAR or VARCHAR type, if your column data fits in maximum CHAR size (254) or VARCHAR size (32768), respectively. For larger column data size, String type should be used as we store its data in CLOB format internally.
Create a table with columns of CHAR and VARCHAR datatype using SQL:
CREATE TABLE tableName (Col1 char(25), Col2 varchar(100)) using row;Create a table with columns of CHAR and VARCHAR datatype using API:
import org.apache.spark.sql.collection.Utils
import org.apache.spark.sql.types.{IntegerType, StringType, StructField, StructType}
val snappy: SnappySession = new SnappySession(spark.sparkContext)
// define schema for table
val varcharSize = 100
val charSize = 25
val schema = StructType(Array(
StructField("col_varchar", StringType, false, Utils.varcharMetadata(varcharSize)),
StructField("col_char", StringType, false, Utils.charMetadata(charSize))
))
// create the table
snappy.createTable(tableName, "row", schema, Map.empty[String, String])Note: STRING columns are handled differently when queried over a JDBC connection.
To ensure optimal performance for SELECT queries executed over JDBC connection (more specifically, those that get routed to lead node), the data of STRING columns is returned in VARCHAR format, by default. This also helps the data visualization tools to render the data effectively.
However, if the STRING column size is larger than VARCHAR limit (32768), you can enforce the returned data format to be in CLOB in following ways:
Using the system property spark-string-as-clob when starting the lead node(s). This applies to all the STRING columns in all the tables in cluster.
bin/snappy leader start -locators:localhost:10334 -J-Dspark-string-as-clob=true
Defining the column(s) itself as CLOB, either using SQL or API. In the example below, we define the column 'Col2' to be CLOB.
CREATE TABLE tableName (Col1 INT, Col2 CLOB, Col3 STRING, Col4 STRING);
import org.apache.spark.sql.collection.Utils
import org.apache.spark.sql.types.{StringType, StructField, StructType}
val snappy: SnappySession = new SnappySession(spark.sparkContext)
// Define schema for table
val schema = StructType(Array(
// The parameter Utils.stringMetadata() ensures that this column is rendered as CLOB
StructField("Col2", StringType, false, Utils.stringMetadata())
))
snappy.createTable(tableName, "column", schema, Map.empty[String, String])Using the query-hint `columnsAsClob in the SELECT query.
SELECT * FROM tableName --+ columnsAsClob(*)
The usage of * above causes all the STRING columns in the table to be rendered as CLOB. You can also provide comma-separated specific column name(s) instead of * above so that data of only those column(s) is returned as CLOB.
SELECT * FROM tableName --+ columnsAsClob(Col3,Col4)
Generally, the column table is used for analytical purpose. To this end, most of the operations (read or write) on it are bulk operations. Taking advantage of this fact the rows are compressed column wise and stored.
In SnappyData, the column table consists of two components, delta row buffer and column store. We try to support individual insert of single row, we store them in a delta row buffer which is write optimized and highly available. Once the size of buffer reaches the COLUMN_BATCH_SIZE set by the user, the delta row buffer is compressed column wise and stored in the column store. Any query on column table also takes into account the row cached buffer. By doing this, we ensure that the query does not miss any data.
We use a persistent Hive catalog for all our metadata storage. All table, schema definition are stored here in a reliable manner. As we intend be able to quickly recover from driver failover, we chose GemFireXd itself to store meta information. This gives us the ability to query underlying GemFireXD to reconstruct the meta store in case of a driver failover.
Refer to the SQL Reference Guide for information on the syntax.
SnappyData’s streaming functionality builds on top of Spark Streaming and primarily is aimed at making it simpler to build streaming applications and integration with the built-in store. Here is a brief overview of Spark streaming from the Spark Streaming guide.
###Spark Streaming Overview
Spark Streaming is an extension of the core Spark API that enables scalable, high-throughput, fault-tolerant stream processing of live data streams. Data can be ingested from many sources like Kafka, Flume, Twitter, ZeroMQ, Kinesis, or TCP sockets, and can be processed using complex algorithms expressed with high-level functions like map, reduce, join and window.
Finally, processed data can be pushed out to filesystems, databases, and live dashboards. In fact, you can apply Spark's machine learning and graph processing algorithms on data streams.
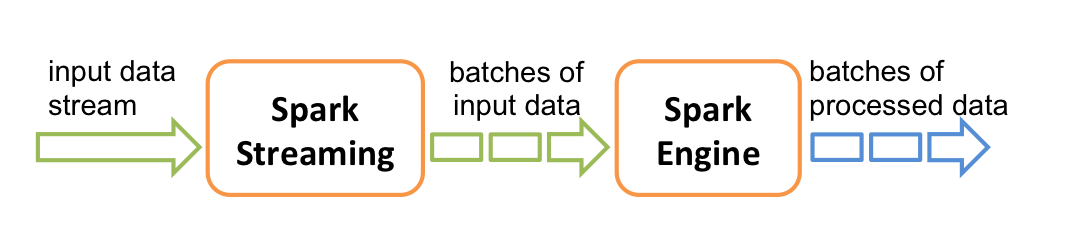
Internally, it works as follows. Spark Streaming receives live input data streams and divides the data into batches, which are then processed by the Spark engine to generate the final stream of results in batches.
Spark Streaming provides a high-level abstraction called discretized stream or DStream, which represents a continuous stream of data. DStreams can be created either from input data streams from sources such as Kafka, Flume, and Kinesis, or by applying high-level operations on other DStreams. Internally, a DStream is represented as a sequence of RDDs.
Additional details on the Spark Streaming concepts and programming is covered here.
We offer the following enhancements over Spark Streaming:
-
Manage Streams declaratively: Similar to SQL Tables, Streams can be defined declaratively from any SQL client and managed as Tables in the persistent system catalog of SnappyStore. The declarative language follows the SQL language and provides access to any of the Spark Streaming streaming adapters such as Kafka or file input streams. Raw tuples arriving can be transformed into a proper structure through pluggable transformers providing the desired flexibility for custom filtering or type conversions.
-
SQL based stream processing: With streams visible as Tables they can be joined with other streams or resident tables (reference data, history, etc). Essentially, the entire SQL language can be used to analyze distributed streams.
-
Continuous queries and time windows: Similar to popular stream processing products, applications can register “continuous” queries on streams. By default, Spark streaming emits batches once every second and any registered queries would be executed each time a batch is emitted. To support arbitrary time ranges, we extend the standard SQL to be able to specify the time window for the query.
-
OLAP optimizations: By integrating and collocating stream processing with our hybrid in-memory storage engine, we leverage our optimizer and column store for expensive scans and aggregations, while providing fast key-based operations with our row store.
-
Approximate stream analytics: When the volumes are too high, a stream can be summarized using various forms of samples and sketches to enable fast time series analytics. This is particularly useful when applications are interested in trending patterns, for instance, rendering a set of trend lines in real time on user displays.
SnappyData supports creation of stream tables from Twitter, Kafka, Files, Sockets sources.
// DDL for creating a stream table
CREATE STREAM TABLE [IF NOT EXISTS] table_name
(COLUMN_DEFINITION)
USING 'kafka_stream | file_stream | twitter_stream | socket_stream | directkafka_stream'
OPTIONS (
// multiple stream source specific options
storagelevel '',
rowConverter '',
topics '',
kafkaParams '',
consumerKey '',
consumerSecret '',
accessToken '',
accessTokenSecret '',
hostname '',
port '',
directory ''
)
// DDL for dropping a stream table
DROP TABLE [IF EXISTS] table_name
// Initialize StreamingContext
STREAMING INIT <batchInterval> [SECS|SECOND|MILLIS|MILLISECOND|MINS|MINUTE]
// Start streaming
STREAMING START
// Stop streaming
STREAMING STOPFor example to create a stream table using kafka source :
val spark: SparkSession = SparkSession
.builder
.appName("SparkApp")
.master("local[4]")
.getOrCreate
val snsc = new SnappyStreamingContext(spark.sparkContext, Duration(1))
snsc.sql("create stream table streamTable (userId string, clickStreamLog string) " +
"using kafka_stream options (" +
"storagelevel 'MEMORY_AND_DISK_SER_2', " +
"rowConverter 'io.snappydata.app.streaming.KafkaStreamToRowsConverter', " +
"kafkaParams 'zookeeper.connect->localhost:2181;auto.offset.reset->smallest;group.id->myGroupId', " +
"topics 'streamTopic:01')")
// You can get a handle of underlying DStream of the table
val dStream = snsc.getSchemaDStream("streamTable")
// You can also save the DataFrames to an external table
dStream.foreachDataFrame(_.write.insertInto(tableName))The streamTable created in the above example can be accessed from snappy-sql and can be queried using ad-hoc SQL queries.
Start a SnappyData cluster and connect through snappy-sql :
//create a connection
snappy> connect client 'localhost:1527';
// Initialize streaming with batchInterval of 2 seconds
snappy> streaming init 2secs;
// Create a stream table
snappy> create stream table streamTable (id long, text string, fullName string, country string,
retweets int, hashtag string) using twitter_stream options (consumerKey '', consumerSecret '',
accessToken '', accessTokenSecret '', rowConverter 'org.apache.spark.sql.streaming.TweetToRowsConverter');
// Start the streaming
snappy> streaming start;
//Run ad-hoc queries on the streamTable on current batch of data
snappy> select id, text, fullName from streamTable where text like '%snappy%'
// Drop the streamTable
snappy> drop table streamTable;
// Stop the streaming
snappy> streaming stop;SchemaDStream is SQL based DStream with support for schema/Product. It offers the ability to manipulate SQL queries on DStreams. It is similar to SchemaRDD, which offers similar functions. Internally, RDD of each batch duration is treated as a small table and CQs are evaluated on those small tables. Similar to foreachRDD in DStream, SchemaDStream provides foreachDataFrame API. SchemaDStream can be registered as a table. Some of these ideas (especially naming our abstractions) were borrowed from Intel's Streaming SQL project.
//You can join two stream tables and produce a result stream.
val resultStream = snsc.registerCQ("SELECT s1.id, s1.text FROM stream1 window (duration
'2' seconds, slide '2' seconds) s1 JOIN stream2 s2 ON s1.id = s2.id")
// You can also save the DataFrames to an external table
dStream.foreachDataFrame(_.write.insertInto("yourTableName"))Unlike Spark streaming, you do not need to register all your stream output transformations (which is a continuous query in this case) before the start of StreamingContext. The continuous queries can be registered even after the SnappyStreamingContext has started.
Users can define a function and completely customize how SnappyData evaluates data and manipulates queries using UDF and UDAF functions across sessions. The definition of the functions is stored in a persistent catalog, which enables it to be used after node restart as well.
Note: Support for UDFs is available in SnappyData 0.8 and higher.
You can simply extend any one of the interfaces in the package org.apache.spark.sql.api.java. These interfaces can be included in your client application by adding snappy-spark-sql_2.11-2.0.3-2.jar to your classpath.
The number in the interfaces (UDF1 to UDF22) signifies the number of parameters an UDF can take.
Note: Currently, any UDF which can take more than 22 parameters is not supported.
package some.package
import org.apache.spark.sql.api.java.UDF1
class StringLengthUDF extends UDF1[String, Int] {
override def call(t1: String): Int = t1.length
}
After defining an UDF you can bundle the UDF class in a JAR file and create the function by using ./bin/snappy-shell of SnappyData. This creates a persistent entry in the catalog after which, you use the UDF.
CREATE FUNCTION udf_name AS qualified_class_name RETURNS data_type USING JAR '/path/to/file/udf.jar'
For example:
CREATE FUNCTION APP.strnglen AS some.package.StringLengthUDF RETURNS Integer USING JAR '/path/to/file/udf.jar'
[Rishi] we can modify the declaration section as above
You can write a JAVA or SCALA class to write an UDF implementation.
Note: For input/output types:
The framework always returns the Java types to the UDFs. So, if you are writing scala.math.BigDecimal as an input type or output type, an exception is reported. You can use java.math.BigDecimal in the SCALA code.
**Return Types to UDF program type mapping **
| SnappyData Type | UDF Type |
|---|---|
| STRING | java.lang.String |
| INTEGER | java.lang.Integer |
| LONG | java.lang.Long |
| DOUBLE | java.lang.Double |
| DECIMAL | java.math.BigDecimal |
| DATE | java.sql.Date |
| TIMESTAMP | java.sql.Timestamp |
| FLOAT | java.lang.Float |
| BOOLEAN | java.lang.Boolean |
| SHORT | java.lang.Short |
| BYTE | java.lang.Byte |
select strnglen(string_column) from <table>
If you try to use an UDF on a different type of column, for example, an Int column an exception is reported.
DROP FUNCTION IF EXISTS udf_name
For example:
DROP FUNCTION IF EXISTS app.strnglen
-
Drop the existing UDF
-
Modify the UDF code and create a new UDF. You can create the UDF with the same name as that of the dropped UDF.
SnappyData uses same interface as that of Spark to define a User Defined Aggregate Function org.apache.spark.sql.expressions.UserDefinedAggregateFunction. For more information refer to this document.