SCGV is an interactive graphical tool for single-cell genomics data, with emphasis on single-cell genomics of cancer. It facilitates examination, jointly or individually, of DNA copy number profiles of cells harvested from multiple anatomic locations (sectors). In the opening view the copy-number data matrix, with columns corresponding to cells and rows to genomic locations, is represented as a heat map with color-encoded integer DNA copy number. If a phylogenetic tree is available for the cells comprising the dataset, it can be used to order the columns of the data matrix, and clones formed by closely related cells may be identified. Alternatively, the columns can be ordered by the sector of origin of the cells. Cyto-pathological information may be displayed in a separate view, including sector-specific slide images and pathology reports. Genomic sub-regions and random subsets of cells can be selected and zoomed into. Individual or multiple copy-number profiles may be plotted as copy number against the genomic coordinate, and these plots may again be zoomed into. Chromosomal regions selected within the profiles may be followed to UCSC genome browser to examine the genomic context.
Short video introduction to SCGV can be found here:
-
Go to anaconda web site https://www.anaconda.com/distribution/ and download the latest anaconda installer for your operating system. SCGV uses Python 3.6 or later so you need to choose the appropriate Anaconda installer.
-
Install anaconda into suitable place on your local machine following instructions from https://docs.anaconda.com/anaconda/install/
-
To create
scgvenvironment use:conda create -n scgv
-
Activate the newly created environment with:
conda activate scgv
-
To install SCGV you can use KrasnitzLab Anaconda channel:
conda install -c krasnitzlab scgv
-
To run the SCGV viewer run following command:
scgview
-
Files in the dataset should conform to the following naming convention. Each filename should end with two dot-separated words. The last word is the usual file extension and second to last is the file type. For example:
example.featuremat.txt
is a
txtfile, that containsfeaturematused by the viewer. -
Example dataset directory is located in subdirectory
exampledata/example.directoryof the project main directory. The content of the example dataset directory is as follows:. ├── example.cells.csv ├── example.clone.txt ├── example.genome.txt ├── example.guide.txt ├── example.featuremat.txt ├── example.features.txt ├── example.ratio.txt ├── example.seg.txt ├── example.tree.txt └── pathology ├── 9727420.color.map.060414.png ├── Area1.Benign.jpg ├── Area1.Benign.txt ├── Area2.PIN.with.Benign.jpg ├── Area2.PIN.with.Benign.txt ├── Area3.GS9.invading.SV.jpg ├── Area3.GS9.invading.SV.txt ├── Area4.GS9.near.Urethra.jpg ├── Area4.GS9.near.Urethra.txt ├── Area5.GS9.at.Capsule.jpg ├── Area5.GS9.at.Capsule.txt └── description.csv -
Optionally the dataset directory may contain a
pathologysubdirectory that contains pathology images and notes. This subdirectory should contain a file calleddescription.csvwith the following structure:sector,pathology,image,notes 1,Benign prostatic tissue,Area1.Benign.jpg,Area1.Benign.txt 2,Pin and benign prostate,Area2.PIN.with.Benign.jpg,Area2.PIN.with.Benign.txt 3,Gleason 9 and invading seminal vesicle,Area3.GS9.invading.SV.jpg,Area3.GS9.invading.SV.txt 4,Gleason 9 near urethra,Area4.GS9.near.Urethra.jpg,Area4.GS9.near.Urethra.txt 5,Gleason 9 at capsule,Area5.GS9.at.Capsule.jpg,Area5.GS9.at.Capsule.txt
First column in description.csv contains the name/id of the sector as
specified in the guide file,
the second column is a description of the sector and the last two columns
contain file names of pathology image and notes.
-
Viewer supports datasets stored as a
ZIParchives. Files from the archive dataset should follow the same naming convention as for dataset directories. -
Example dataset ZIP archive is found in
exampledata/example.archive.zipinto project main directory. The structure of the example dataset is as follows:unzip -t example.archive.zip Archive: example.archive.zip testing: pathology/ OK testing: pathology/Area5.GS9.at.Capsule.jpg OK testing: pathology/Area4.GS9.near.Urethra.jpg OK testing: pathology/Area3.GS9.invading.SV.jpg OK testing: pathology/Area2.PIN.with.Benign.jpg OK testing: pathology/Area1.Benign.jpg OK testing: pathology/Area5.GS9.at.Capsule.txt OK testing: pathology/Area4.GS9.near.Urethra.txt OK testing: pathology/Area1.Benign.txt OK testing: pathology/description.csv OK testing: pathology/Area2.PIN.with.Benign.txt OK testing: pathology/Area3.GS9.invading.SV.txt OK testing: example.cells.csv OK testing: example.clone.txt OK testing: example.featuremat.txt OK testing: example.features.txt OK testing: example.genome.txt OK testing: example.guide.txt OK testing: example.ratio.txt OK testing: example.seg.txt OK testing: example.tree.txt OK No errors detected in compressed data of example.archive.zip.
-
Before starting the viewer you need to activate viewer's Anaconda environment
conda activate scgv
-
To start the viewer use:
scgview
-
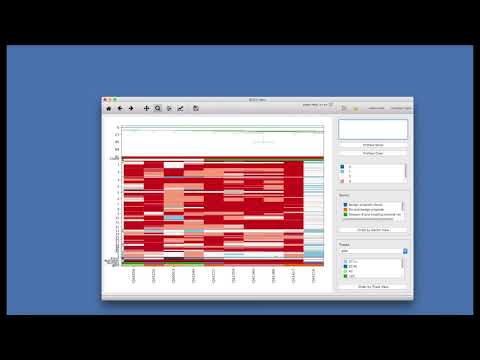
The startup window of SCGV should appear:
-
Use
Open Directory(1) andOpen Archive(2) buttons to open a data set for visualization -
Open Directorybutton allows you to select a directory where a dataset is located. One directory may contain only one dataset. -
Open Archivebutton allow you to select dataset stored as aZIParchive.
-
After dataset is loaded it will displayed into the main window.

-
From profiles instruments you can select individual cells to display their CN profile into single profile viewer.
-
Buttons 'Feature View' and 'Order by Sector View' will display different views of the whole dataset
-
From 'Sectors' legend you can visualize single sector view and pathology view for any given sector.
-
If you right click on a single cell it will be added to list of profiles to visualize from 'Show Profiles' button.
-
To show the selected profiles you need to click on 'Profiles Show' button. Selected profiles will be visualized as stacked plot of copy number against the genomic coordinate:
 |
|---|
| 1-reset view; 2-undo; 3-redo; 4-move the image; 5-zoom in; 6-save an image snapshot; 7,8-individual profiles stacked view |
- Using zoom button (5) and reset button (1) you can inspect different regions of the profile.
- To examine the genomic content of an intra-chromosomal region, right-click and use the context menu to select desired genomic position or region in the stacked copy-number profile view. This will invoke UCSC Genome Browser in a tab of your default web browser
-
Right click on a sector in Sector legeng will open a context menu:
-
Selecting 'Show sector view' displays single sector view for the selected sector
-
Selecting 'Show sector patology' will display pathology image and notes for the selected sector
Using 'Configure Tracks' (6) button you can configure tracks to be shown on bottom of the heatmap view.
-
When you click 'Configure Tracks' (6) button a dialog window is shown allowing you to select additional tracks to be displyed
-
Once selected the configured tracks are shown at the bottom of the heatmap view and are available from 'Track legend selector' (11):
-
The 'Selected track legend' (12) allows you additionally explore the selected track using context menu: