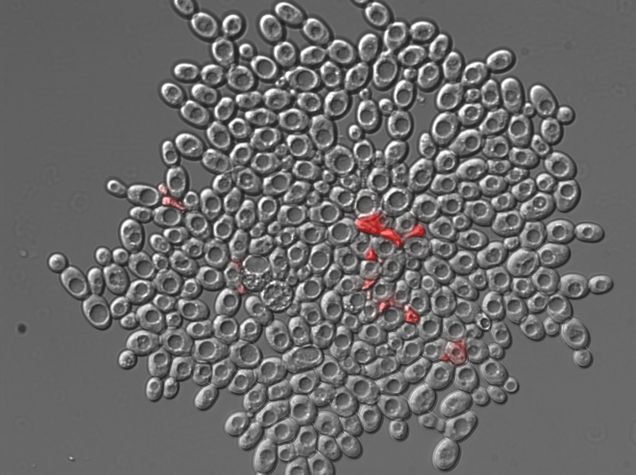
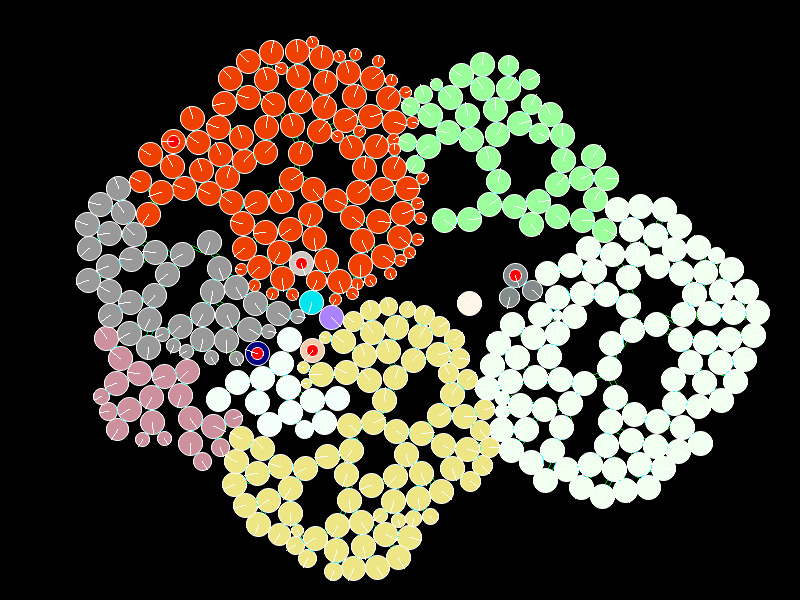
The yeast Saccharomyces cerevisiae can evolve a simple form of multicellularity in lab conditions which gravity-select for larger clumps (1,2). An analysis of this growth suggests that the geometry of this growth has important evolutionary consequences, and can explain why an initial division of reproductive labour arises (3). The division of labour exhibits itself as an increased cell-death rate within the clump. Dead cells provide weak points, allowing the clusters to split, increasing the space for further replication. So increased cell death actually increases the overall reproductive rate of cells and clusters.
This aim of this simulation is to provide a simple in-silico visual laboratory for exploring the parameters that control cluster growth and splitting (there is no selection in this model, yet). The simulation uses simple physics to model two-dimensional cluster growth, and provides an optional visual display. Configuration files can provide different sets of parameters, multiple replicates can be run, and the results can be recorded for analysis. This is a work in progress, and more work is needed to capture some of the more interesting dynamics.
You can see what the simulations look like on YouTube, here, and here.
To run this code, you'll need to install pymunk. To see the visualisations, you'll need to install pygame. The simulations will work fine without pygame (indeed, things will run faster), but you don't get any visual feedback.
To run a simulation, you need to invoke python run_simulation.py, and point it at a configuration file. Some example configuration files are in the ./experiments folder. Here is the contents of simple.cfg:
p = parameters()
p.max_steps = 2000
p.max_joint_distortion = 30
p.max_dead_joint_distortion = 5
p.life_expectancy = 5000
p.max_cells = 400
add_treatment('simple', p, replicates=10)This runs 10 simulations with the parameter settings shown. To run this:
$ python run_simulation experiments/simple.cfg
If you want to see some graphics, add the -g flag:
$ python run_simulation -g experiments/simple.cfg
To control the frequency of updates to the graphical output, use the -u <N> option:
$ python run_simulation -g -u 10 experiments/simple.cfg
If you invoke the graphics, you can hit <space> to pause the simulation, and <enter> to take a snapshot of the simulation.
To really play with this simulation, and write your own experiments, you're going to have to dig into the code. The ./yeasty/parameters.py file is a good place to start. The Parameters class provides a bunch of defaults, which can be overridden. Here is a snippet of that file:
start_size = 5
finish_size = 25
grow_step = .1
linear_damping = 0
single_branch_prob = .3
wait_till_grow = 100
max_joint_distortion = 30
max_dead_joint_distortion = 10
max_cells = 400
max_steps = 2000
check_break_every = 1
life_expectancy = 15000A configuration file allows you to create different "treatments" which vary these parameters, and this runs multiple related simulations from one configuration file. To collect the results, you need to load an "analysis" plug-in. A few examples are currently provided, such as one that records the final number of clusters produced at the end of each simulation in a .csv file.
I'm a philosopher of science and a programmer, interested in the evolution of complexity. My website is here.
The programming was done in python by Brett Calcott, in consultation with Ben Kerr. The simulation uses pymunk, the python version of the 2d physics library chipmunk, and the visualisations use pygame. The photo of the yeast is by William C. Ratcliff from the University of Minnesota.