-
Notifications
You must be signed in to change notification settings - Fork 7
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
1 parent
ee67b6b
commit 8ba2bbc
Showing
7 changed files
with
297 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -685,6 +685,9 @@ | |
| "1.1.1", | ||
| "1.1.0" | ||
| ], | ||
| "MMV-playground": [ | ||
| "0.1.3" | ||
| ], | ||
| "morphometrics": [ | ||
| "0.0.8", | ||
| "0.0.7", | ||
|
|
||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,105 @@ | ||
| { | ||
| "name": "MMV-playground", | ||
| "display_name": "MMV-playground", | ||
| "visibility": "public", | ||
| "icon": "", | ||
| "categories": [ | ||
| "Annotation", | ||
| "Segmentation", | ||
| "Acquisition" | ||
| ], | ||
| "schema_version": "0.2.1", | ||
| "on_activate": null, | ||
| "on_deactivate": null, | ||
| "contributions": { | ||
| "commands": [ | ||
| { | ||
| "id": "MMV-playground.make_mmv_playground", | ||
| "title": "Make MMV playground", | ||
| "python_name": "mmv_playground._widget:mmv_playground", | ||
| "short_title": null, | ||
| "category": null, | ||
| "icon": null, | ||
| "enablement": null | ||
| } | ||
| ], | ||
| "readers": null, | ||
| "writers": null, | ||
| "widgets": [ | ||
| { | ||
| "command": "MMV-playground.make_mmv_playground", | ||
| "display_name": "MMV-playground", | ||
| "autogenerate": false | ||
| } | ||
| ], | ||
| "sample_data": null, | ||
| "themes": null, | ||
| "menus": {}, | ||
| "submenus": null, | ||
| "keybindings": null, | ||
| "configuration": [] | ||
| }, | ||
| "package_metadata": { | ||
| "metadata_version": "2.2", | ||
| "name": "MMV-playground", | ||
| "version": "0.1.3", | ||
| "dynamic": null, | ||
| "platform": null, | ||
| "supported_platform": null, | ||
| "summary": "Napari plugin for 2D microscopy segmentation", | ||
| "description": "# MMV-playground\n\n[](https://github.com/MMV-Lab/MMV-playground/raw/main/LICENSE)\n[](https://pypi.org/project/MMV-playground)\n[](https://python.org)\n[](https://github.com/MMV-Lab/MMV-playground/actions)\n[](https://codecov.io/gh/MMV-Lab/MMV-playground)\n[](https://napari-hub.org/plugins/MMV-playground)\n\nThis plugin is aimed at researchers in biology and medicine who want to segment and analyze 2D microscopy images. It offers intuitive tools for common pre-processing and analysis tasks.\n\n----------------------------------\n\nThis [napari] plugin was generated with [Cookiecutter] using [@napari]'s [cookiecutter-napari-plugin] template.\n\n<!--\nDon't miss the full getting started guide to set up your new package:\nhttps://github.com/napari/cookiecutter-napari-plugin#getting-started\n\nand review the napari docs for plugin developers:\nhttps://napari.org/stable/plugins/index.html\n-->\n\n## Installation\n\nYou can install `MMV-playground` via [pip]:\n\n pip install MMV-playground\n\nTo install latest development version :\n\n pip install git+https://github.com/MMV-Lab/MMV-playground.git\n\n## Documentation\n\nThis plugin for the graphics software Napari is designed to analyze two-dimensional microscopy images. Images should be provided in grayscale format, as colored images are not supported. The plugin includes seven core functions for image analysis:\n\n1. Intensity normalization \n2. Smoothing \n3. Background correction \n4. Spot-shape filter \n5. Filament-shape filter \n6. Thresholding \n7. Topology-preserving thinning \n\n### **How to Start and Use the Plugin**\n\nTo start the plugin, open Napari, go to the \"Plugins\" menu, and select \"MMV-playground (MMV-playground).\" The MMV-playground interface will appear on the right-hand side of the Napari window, displaying seven buttons, each corresponding to one of the available functions. Clicking a button opens a dialog box where you can select an image, adjust the parameters for the chosen function, and execute it by pressing the \"Run\" button. Clicking the function button again collapses the dialog box.\n\n### Screenshot\n\nHere is a preview of the MMV-playground plugin in action:\n\n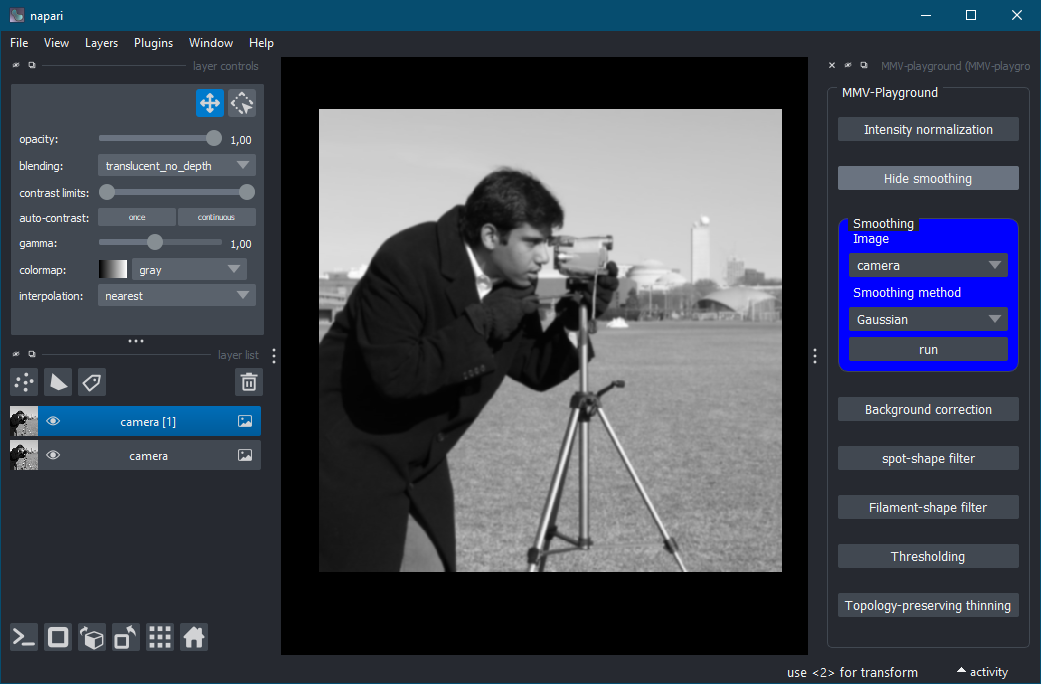\n\n---\n\n### **Intensity Normalization**\n\nThis function adjusts the image intensity to enhance contrast and improve uniformity. Two percentage values are required: \n- *Lower percentage (0\u20135%)*: Defines the darkest portion of the image to ignore as background noise, which is set to a fixed value. \n- *Upper percentage (95\u2013100%)*: Specifies the brightest portion of the image to be capped, preventing overexposure. \n\nThe plugin calculates the respective percentiles based on these values. Intensities below the lower percentile are clipped, and those above the upper percentile are also capped. Finally, all pixel intensities are rescaled to the range [0, 1].\n\n---\n\n### **Smoothing**\n\nThis function reduces noise to enhance image clarity. Two methods are available: \n- *Gaussian smoothing* \n- *Edge-preserving smoothing*: Retains edges (e.g., cell boundaries) while reducing noise. \n\n---\n\n### **Background Correction**\n\nThis function removes uneven illumination or background artifacts using a filter. The key parameter is: \n- *Kernel size (1\u2013100)*: Determines the spatial scale of the background correction. Smaller kernel sizes remove local noise, while larger sizes correct for broader illumination variations. \n\nThe [scipy.ndimage.white_tophat] function is used to perform the correction, making this method effective for images with dark backgrounds.\n\n---\n\n### **Spot-Shape Filter**\n\nThis filter detects circular structures, such as cell nuclei or fluorescent spots. It is based on the [scipy.ndimage.gaussian_laplace] function and requires: \n- *Sigma (\u03c3)*: Controls the size of the spots to detect. Smaller sigma values target smaller spots, while larger values focus on larger structures.\n\n---\n\n### **Filament-Shape Filter**\n\nThis filter highlights elongated structures like cytoskeletal fibers or blood vessels. Using the [aicssegmentation.core.vessel.vesselness2D] function, the key parameter is: \n- *Sigma (\u03c3)*: Specifies the width of the detected filaments. Lower values detect thinner structures, while higher values identify thicker ones.\n\n---\n\n### **Thresholding**\n\nThis function segments the image into binary regions by separating the signal from the background. Users can choose one of four thresholding methods: \n- *Otsu*: Best for images with clear separation between background and signal intensities. \n- *Li*: Suitable for uniformly illuminated images. \n- *Triangle*: Effective for asymmetrical intensity distributions. \n- *Sauvola*: Ideal for images with uneven illumination. \n\nThe result is a binary image where pixels above the threshold are set to 1 (signal), and all others are set to 0 (background).\n\n---\n\n### **Topology-Preserving Thinning**\n\nThis function extracts the skeleton of structures while maintaining their connectivity. Two parameters are required: \n- *Minimum thickness (0.5\u20135)*: Defines the smallest allowable thickness of structures before thinning. \n- *Thin (1\u20135)*: Controls the degree of thinning, reducing structure width while preserving topology (e.g., network connections). \n\nThis is particularly useful for analyzing vascular or cellular networks.\n\n---\n\n### **What Is Missing?**\n\nCurrently, unit tests are not implemented, and internal documentation of the source code is still in progress.\n\n## Contributing\n\nContributions are very welcome. Tests can be run with [tox], please ensure\nthe coverage at least stays the same before you submit a pull request.\n\n## License\n\nDistributed under the terms of the [BSD-3] license,\n\"MMV-playground\" is free and open source software\n\n## Issues\n\nIf you encounter any problems, please [file an issue] along with a detailed description.\n\n[napari]: https://github.com/napari/napari\n[Cookiecutter]: https://github.com/audreyr/cookiecutter\n[@napari]: https://github.com/napari\n[MIT]: http://opensource.org/licenses/MIT\n[BSD-3]: http://opensource.org/licenses/BSD-3-Clause\n[GNU GPL v3.0]: http://www.gnu.org/licenses/gpl-3.0.txt\n[GNU LGPL v3.0]: http://www.gnu.org/licenses/lgpl-3.0.txt\n[Apache Software License 2.0]: http://www.apache.org/licenses/LICENSE-2.0\n[Mozilla Public License 2.0]: https://www.mozilla.org/media/MPL/2.0/index.txt\n[cookiecutter-napari-plugin]: https://github.com/napari/cookiecutter-napari-plugin\n\n[file an issue]: https://github.com/MMV-Lab/MMV-playground/issues\n\n[napari]: https://github.com/napari/napari\n[tox]: https://tox.readthedocs.io/en/latest/\n[pip]: https://pypi.org/project/pip/\n[PyPI]: https://pypi.org/\n\n[scipy.ndimage.gaussian_filter]: https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.gaussian_filter.html\n[itk.GradientAnisotropicDiffusionImageFilter]: https://itk.org/Doxygen/html/classitk_1_1GradientAnisotropicDiffusionImageFilter.html\n[scipy.ndimage.white_tophat]: https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.white_tophat.html\n[scipy.ndimage.gaussian_laplace]: https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.gaussian_laplace.html\n[aicssegmentation.core.vessel.vesselness2D]: https://allencell.github.io/aics-segmentation/aicssegmentation.core.html\n", | ||
| "description_content_type": "text/markdown", | ||
| "keywords": null, | ||
| "home_page": null, | ||
| "download_url": null, | ||
| "author": "Peter Lampen", | ||
| "author_email": "[email protected]", | ||
| "maintainer": null, | ||
| "maintainer_email": null, | ||
| "license": "Copyright (c) 2024, Peter Lampen\nAll rights reserved.\n\nRedistribution and use in source and binary forms, with or without\nmodification, are permitted provided that the following conditions are met:\n\n* Redistributions of source code must retain the above copyright notice, this\n list of conditions and the following disclaimer.\n\n* Redistributions in binary form must reproduce the above copyright notice,\n this list of conditions and the following disclaimer in the documentation\n and/or other materials provided with the distribution.\n\n* Neither the name of copyright holder nor the names of its\n contributors may be used to endorse or promote products derived from\n this software without specific prior written permission.\n\nTHIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS \"AS IS\"\nAND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE\nIMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE\nDISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE\nFOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL\nDAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR\nSERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER\nCAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,\nOR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE\nOF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.\n", | ||
| "classifier": [ | ||
| "Development Status :: 2 - Pre-Alpha", | ||
| "Framework :: napari", | ||
| "Intended Audience :: Developers", | ||
| "License :: OSI Approved :: BSD License", | ||
| "Operating System :: OS Independent", | ||
| "Programming Language :: Python", | ||
| "Programming Language :: Python :: 3", | ||
| "Programming Language :: Python :: 3 :: Only", | ||
| "Programming Language :: Python :: 3.9", | ||
| "Programming Language :: Python :: 3.10", | ||
| "Programming Language :: Python :: 3.11", | ||
| "Programming Language :: Python :: 3.12", | ||
| "Topic :: Scientific/Engineering :: Image Processing" | ||
| ], | ||
| "requires_dist": [ | ||
| "aicssegmentation", | ||
| "itk", | ||
| "napari", | ||
| "numpy", | ||
| "qtpy", | ||
| "scikit-image", | ||
| "scipy", | ||
| "tox; extra == \"testing\"", | ||
| "pytest; extra == \"testing\"", | ||
| "pytest-cov; extra == \"testing\"", | ||
| "pytest-qt; extra == \"testing\"", | ||
| "napari; extra == \"testing\"", | ||
| "pyqt5; extra == \"testing\"" | ||
| ], | ||
| "requires_python": ">=3.9", | ||
| "requires_external": null, | ||
| "project_url": [ | ||
| "Bug Tracker, https://github.com/MMV-Lab/MMV-playground/issues", | ||
| "Documentation, https://github.com/MMV-Lab/MMV-playground#README.md", | ||
| "Source Code, https://github.com/MMV-Lab/MMV-playground", | ||
| "User Support, https://github.com/MMV-Lab/MMV-playground/issues" | ||
| ], | ||
| "provides_extra": [ | ||
| "testing" | ||
| ], | ||
| "provides_dist": null, | ||
| "obsoletes_dist": null | ||
| }, | ||
| "npe1_shim": false | ||
| } |
Oops, something went wrong.