-
Notifications
You must be signed in to change notification settings - Fork 10
Lab 5: Computational Motif Discovery
This lab is to guide you through the implemention of Naive consensus algorithm and Gibbs sampling algorithm for computational motif detection.
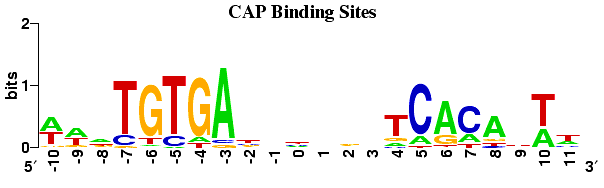
In essence, motif discovery is a pattern discovery problem. This problem can be simplified as follows:
Given a set of DNA sequences, find an unknown pattern that occurs frequently.
A DNA motif is defined as a nucleic acid sequence pattern that has some biological significance. Normally, the pattern is fairly short (5 to 20 bp long) and is known to recur in different genes or several times within a gene.
Algorithms to find regulatory elements can be categorized into two groups:
- (1) methods based on word counting and
- (2) methods based on probabilistic sequence models.
The word counting methods analyze the frequency of oligonucleotides in the upstream regions of the genes of interest and use intelligent strategies to speed up counting and to detect significantly over-represented motifs. These methods then compile a common motif by grouping similar words. Word counting methods lead to a global solution as compared to the probabilistic methods and are appropriate for short motifs and are therefore useful for motif finding in eukaryotic genome where motifs are generally shorter than prokaryotes. These algorithms are a good choice for finding totally constrained motifs, i.e., all instances are identical. However, for typical motifs that often have several weakly conserved positions, word-based methods can be problematic and results often has to be post-processed. In addition, word-based methods also suffer from too many spurious discoveries.
In probabilistic approach, the model parameters are often estimated using Maximum-Likelihood principle or Bayesian inference. Many of the algorithms developed from the probabilistic approach are designed to find longer or more general motifs than are required for transcription factor binding sites. Therefore, they are more suitable for prokaryotic motif discovery. Unfortunately, these algorithms do not guarantee to finding global optima, since some local search methods (e.g. Gibbs sampling, Expectation-Maximization, or greedy algorithms) are employed, which may converge to local optima.
For the
This algorithm finds a position weight matrix (PWM) motif in a set of input DNA sequences.
-
Input:
- a set of sequences
$S={s_1, s_2, \dots, s_n}$ stored in a FASTA file; - the desired motif length,
$k$ - number of motifs to report,
$T$
- a set of sequences
-
Output:
- a set of motifs
$M={m_1, m_2, \dots, m_T}$ , where$m_i$ is$n$ $k$ -mers, each is the subsequence of$s$ .
- a set of motifs
-
Procedure:
- For each pair of
$k$ -mers$x_1$ and$x_2$ from$s_1$ and$s_2$ , compute the information content of the corresponding PWM. Store the$T$ highest PWMs. - For
$i=3,\dots,n$ :- : For every
$k$ -mer$x_i$ (substring of$s_i$ ), add$x_i$ to each PWMs to obtain a new motif, and compute the information content of the corresponding PWM. Store the T highest scoring PWMs.
- : For every
- Output the T highest scoring PWMs as the final motif candidates.
- For each pair of
Procedure:
- Set
$(x_1, \dots, x_n)$ as the random start positions for the motif in the input sequences. That is,$x_i \in [1, |s_i|-k+1]$ -
Repeat until
$(x_1, \dots, x_n)$ not change-
For
$i=1,\dots,n$ - build the profile
$\mathbf{Q}$ with$k$ -mers starting at$(x_1, \dots, x_{i-1}, x_{i+1}, \dots, x_n)$ - update
$x_i$ according to the profile probability distribution of$\mathbf{Q}$ in$s_i$
- build the profile
-
For
-
Output the start sites
$(x_1, \dots, x_n)$
- Implement the above algorithms in C or Python.
- Run your programs on the following toy data, to identify the possible motifs in the set of sequences using the parameters of
$k=9$ and the background base distribution$q_A=q_T=0.35, q_C=q_G=0.15$ :
- (1)
course/bi462/lab5/bicoid.fa - (2)
course/bi462/lab5/hunchback.fa - (3)
course/bi462/lab5/kruppel.fa
- Use the weblogo to obtain the graphical representation of the above motifs.
- What is the difference between the above two algorithms, based on your results?
- Are there any drawbacks for the above approaches? If yes, can you figure out how to improve the algorithm?
- Web-based MEME search of the motifs on the above datasets
- Navigate to MEME-Suite and select MEME from the Motif Discovery section on the left
- Upload your masked sequence file
- Set the number of motifs to be found to 5
- Run the search
- View the HTML output
- Extra exercise
- Download MEME-Suite
- Unpack and Intall the suite, figure out how to use the tool for motif discovery.
- Read the accompanied reference article and also the source code, then comment on how expectation-maximization (EM) algorithm is used to find the motifs in a set of sequences.
On the way to the garden of bioinformatics.
A bioinformatics wiki for the course BI462.