-
Notifications
You must be signed in to change notification settings - Fork 13
AMICA Introduction
AMICA is developed by Jason Palmer (Palmer, ICASSP, 2008, Palmer, Tech Report, 2012). Its applications to EEG data are further validated in Delorme, Plos One, 2012, and Hsu, NeuroImage, 2018. This tutorial is mainly written and maintained by Sheng-Hsiou (Shawn) Hsu with links to Jason's original sources, hoping to provide up-to-date information and detailed instructions on AMICA.
(c) Jason Palmer, University of California San Diego, 2015.
Adaptive Mixture Independent Component Analysis (AMICA) is a binary program (for Linux, Mac, and Windows) that performs an independent component analysis (ICA) decomposition on input data, potentially with multiple ICA models. It can be run standalone, or from MATLAB. Key features of AMICA include:
- Adaptive Source Densities: the source density models are adapted using a mixture of Generalized Gaussian density model, resulting in extremely good fit between the density model and the actual density of the source being estimated
- Multiple / Mixture Models: AMICA allows multiple ICA models to be learned simultaneously, automatically segmenting the data into regions of local stationarity, and returning a set of components for each model. AMICA can also be set to share components between models to increase estimation efficiency
- Data Likelihood (Model Probability): likelihood of each model for given and new data is available, allowing rejection of unlikely data, as well as classification of new data
- Parallel Implementation: program can use multiple cores in single workstations (using portable OpenMP code), as well as multiple nodes in a cluster (using portable MPI code). All binaries allow multi-core (SMP) execution. Only the Linux version currently supports clusters (we use the freely available Rocks / Sun Grid Engine)
For an explanation of how AMICA works, see this page.
- AMICA on Github (contains the latest version)
- AMICA plugin for EEGLAB: direct download (contains the latest stable version)
- AMICA tutorial slides for EEGLAB workshop
- A tutorial introduction to AMICA applied to generated example data.
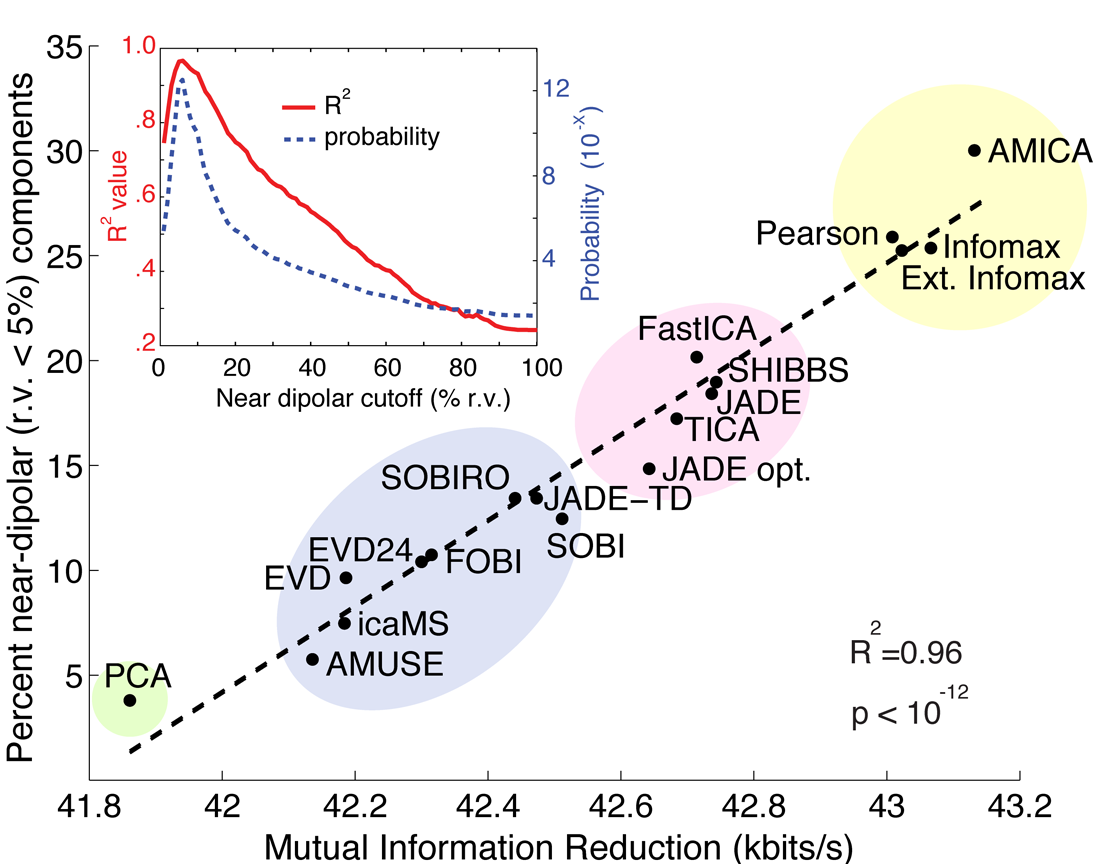
With a complex model that has more parameters to learn, AMICA can achieve an (empirically) more accurate ICA decomposition. Arnauld Delorme et al. in 2012 compared several ICA algorithms and showed that AMICA outperformed all other ICA approaches in terms of obtaining higher mutual information reduction and more near-dipolar ICs.
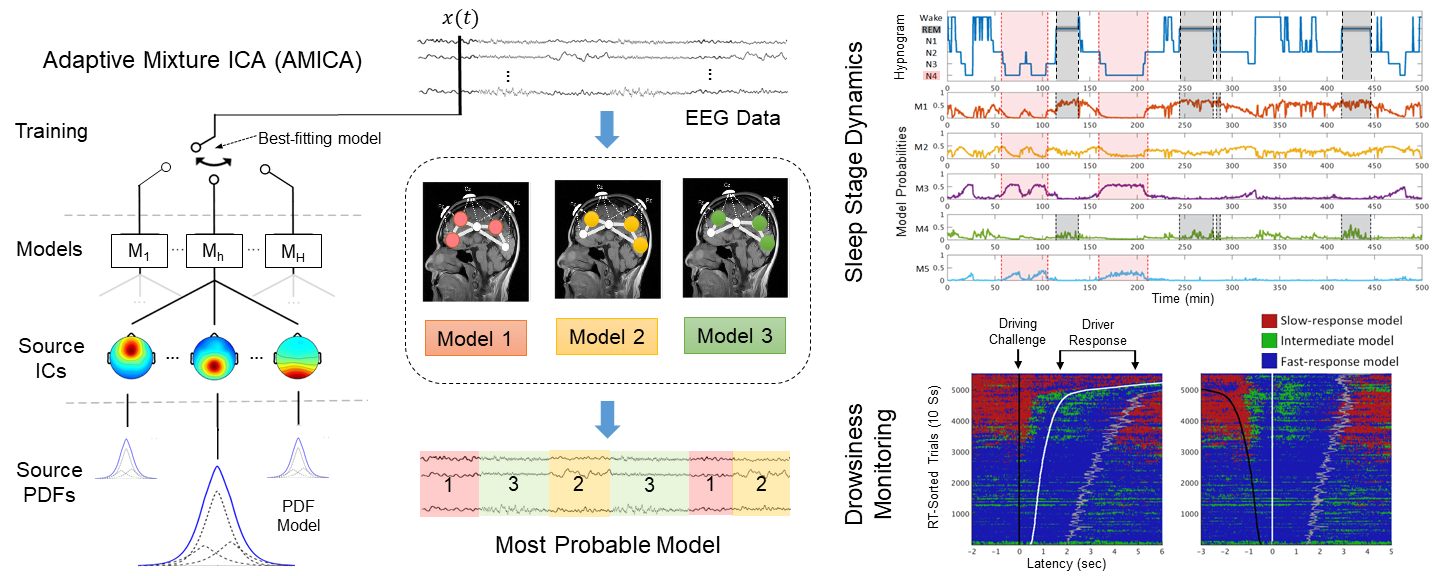
This reason itself already makes AMICA attractive. But AMICA has a capability that has been long-time overlooked – learning a mixture of ICA models.
Hsu and Jung, 2017 hypothesizes that transitions of brain states involve changes in cortical networks activities which can be identified by distinct ICA models. This hypothesis motivates the application of AMICA – with multiple ICA models – as a data-driven approach to modeling nonstationary dynamics of continuous and unlabeled data.
Recent studies (Hsu et al., NeuroImage, 2018 and Hsu et al., BioCAS, 2018) apply multi-model AMICA to modeling EEG dynamics that are associated with brain state changes. We demonstrate that multi-model AMICA can characterize EEG dynamics during sleep for automatic staging, assess transitions between alert and drowsy states in a simulated driving experiment, and uncover mental state changes during a guided-imagery meditation / hypnotherapy. Some empirical results are presented in this section.
We encourage researchers to test the capability of multi-model AMICA in detecting and modeling different cognitive or mental states. For more details, we recommend reading the paper (Hsu, NeuroImage, 2018, PDF)

It is important to point out that AMICA achieves better performance by requesting more computational resources to learn a more complex model and thus requiring longer runtime. Fortunately, AMICA’s inventor, Jason Palmer, derived a Newton-based learning rule and devised a parallel-processing-enabled, pre-compiled program that makes AMICA learning process feasible (e.g. can finish in a few hours instead of a few days).
In 2018, the NIH-funded Neuroscience Gateway (NSG) project aims to provide researchers (free) access to computing power on supercomputer clusters to speed up computation-heavy process, including AMICA. With access to 24 cores per node and potentially to multiple nodes on the supercomputer, the runtime of AMICA can be significantly reduced. More details on how to get access to NSG and how to run AMICA on NSG, please see below.
The figure below gives a schematic overview of the architecture of AMICA and its models.
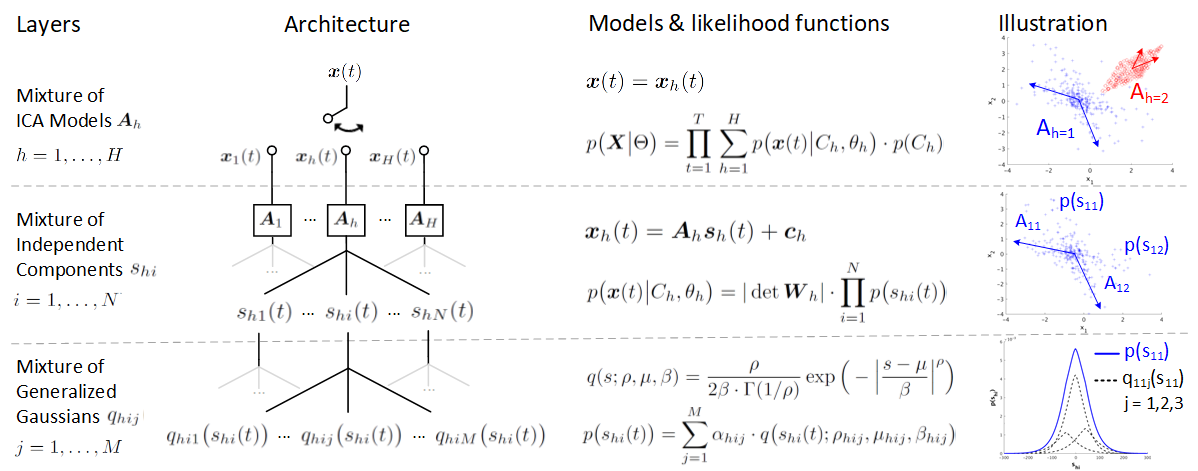
AMICA consists of three layers of mixing:
- First layer: mixture of ICA models \mathbf{A}_1, ... , \mathbf{A}_H that learn the underlying data clusters, as shown in the illustration using simulated data from Laplace and uniform distribution.
- Second layer: mixture of independent components (IC) \mathbf{A}{h1}, ..., \mathbf{A}{hN} that decompose the data cluster into statistically independent sources' activations \mathbf{s}{h1}, ..., \mathbf{s}{hN}.
- Third layer: mixture of generalized Gaussian distribution q(s;\rho,\mu,\beta) that approximate the probability distribution of the source activation p(\mathbf{s})
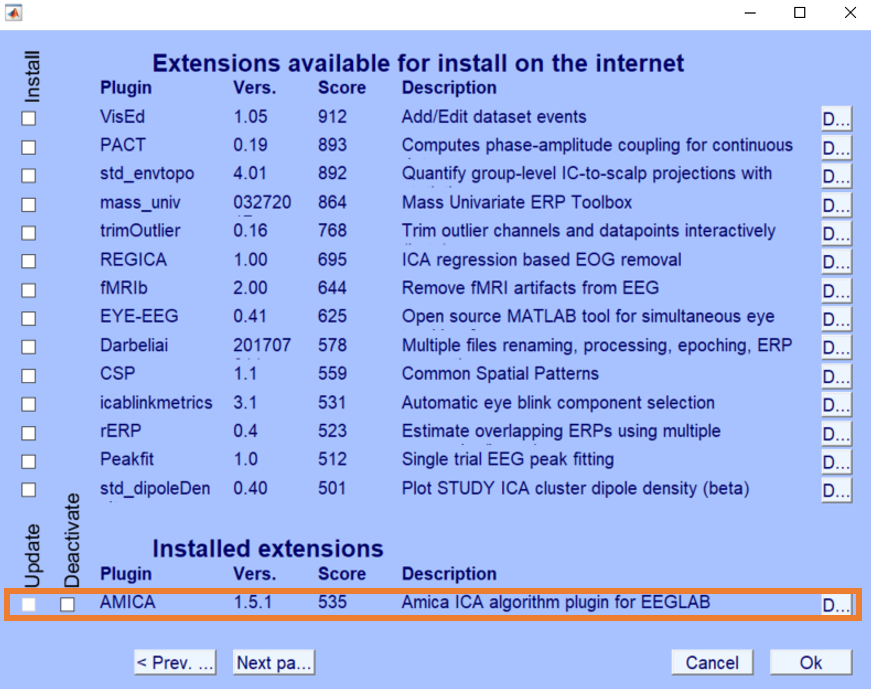
You can download the latest AMICA plugin via the EEGLAB extensions manager from your EEGLAB.
- Open EEGLAB GUI
- Open File --> Manage EEGLAB extensions --> Data processing extensions
- Install Plugin AMICA (Current version 1.5.1, last updated Oct. 2018)
Alternatively, you can download the AMICA plugin folder or fork the AMICA repository on Github directly and put it under the /eeglab/plugins/ folder. More instructions on how to download and install EEGLAB plugins can be found in the EEGLAB Extensions Wiki page.
Note: the following sources may not be up-to-date
- Amica Download on wiki: Amica Download
The downloaded amica1.5.1 package (not amica1.5) should contain all of the compiled binary files for different OS.
- Linux/Unix Users: Plug-and-play! May need to recompile only if runs with multiple nodes or threads.
- Windows Users: Install MPICH2 (32-bit: mpich2-1.4-win-ia32 or 64-bit: mpich2-1.4-win-x86-64), which is included in the amica1.5.1 folder. Otherwise they can be downloaded from http://www.mpich.org/static/downloads/1.4/
- Mac Users: Change the permissions of the binary file amica15mac to executable. Run "Terminal" from the Go->Utilities menu, then at the prompt run:
cd /eeglab/plugins/amica1.5.1
chmod 777 amica15mac
For further information, follow the instruction written by Jason Palmer.
- From EEGLAB GUI
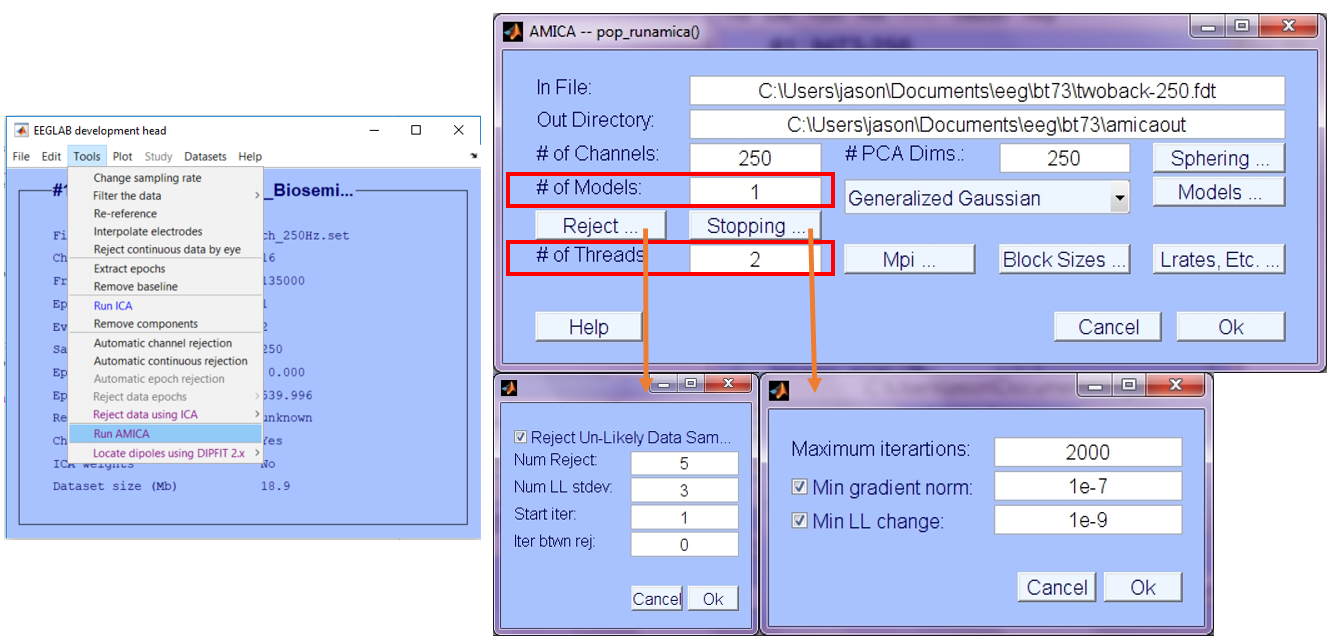
- Tool --> Run AMICA
- Follow the Amica 1.5 EEGLAB GUI Interface Help for setting the input parameters
- Using matlab script: sample script below:
EEG = pop_loadset(‘test_data.set');
% define parameters
numprocs = 1; % # of nodes (default = 1)
max_threads = 1; % # of threads per node
num_models = 1; % # of models of mixture ICA
max_iter = 2000; % max number of learning steps
% run amica
outdir = [ pwd filesep 'amicaouttmp' filesep ];
[weights,sphere,mods] = runamica15(EEG.data, 'num_models',num_models, 'outdir',outdir, ...
'numprocs', numprocs, 'max_threads', max_threads, 'max_iter',max_iter);
% type “help runamica15()” for a full list and explanation of the parameters
EEGLAB scripts may now be run on high-performance computing resources via the freely available Neuroscience Gateway Portal to the NSF-sponsored Comet supercomputer of the San Diego Supercomputer Center. NSG accounts are free and are not limited to US users, but the portal may only be used for non-commercial purposes. You can visit EEGLAB on NSG page for more information.
Computationally expensive operations such as AMICA analysis will be directly benefited with the access to multiple nodes, each with 24 threads / cores, on the Comet supercomputer, which significantly reduces the runtime of AMICA and the expense of local computing resources.
1. Create an NSG account and login: Follow the instructions - EEGLAB on NSG
2. Prepare a MATLAB script that runs AMICA: You can also include EEGLAB preprocessing steps and post-analyses. A sample code below:
%% test_script_amica.m
% add eeglab to path
eeglab; close;
% load dataset
filepath = [ pwd filesep ];
filename = 'test_data.set';
EEG = pop_loadset(filename, filepath);
% define parameters
numprocs = 1; % # of nodes (1-4: default 1)
max_threads = 24; % # of threads (1-24: default = 24)
num_models = 1; % # of models of mixture ICA
max_iter = 2000; % max number of learning steps
% run amica on NSG
outdir = [ pwd filesep 'amicaouttmp' filesep ];
runamica15(EEG.data, 'num_models',num_models, 'outdir',outdir, ...
'numprocs', numprocs, 'max_threads', max_threads, 'max_iter',max_iter);
The runamica15 function will call the binary compiled on Expanse supercomputer that allows for multi-threads and multi-nodes parallel processing. Comprehensive parameters evaluation haven't been completed yet for AMICA on Expanse. But the parameters recommended for the old Comet system still works on Expanse. Recommend set max_threads to 24 since each submitted job will be assigned a node which contains 24 cores on Comet. Requesting more than 1 node is made available by setting numproc larger than 1 (max 4 nodes). Be sure to make the number consistent with the input parameters set while submitting the job on NSG (see step 4).
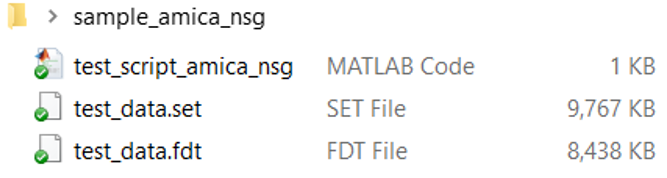 3. Prepare a data folder:
make sure you have the following files in the folder: (1) a Matlab
script, which runs AMICA analysis, and (2) EEG data file(s) called in
the script.
3. Prepare a data folder:
make sure you have the following files in the folder: (1) a Matlab
script, which runs AMICA analysis, and (2) EEG data file(s) called in
the script.
4. Upload the zipped folder to NSG:
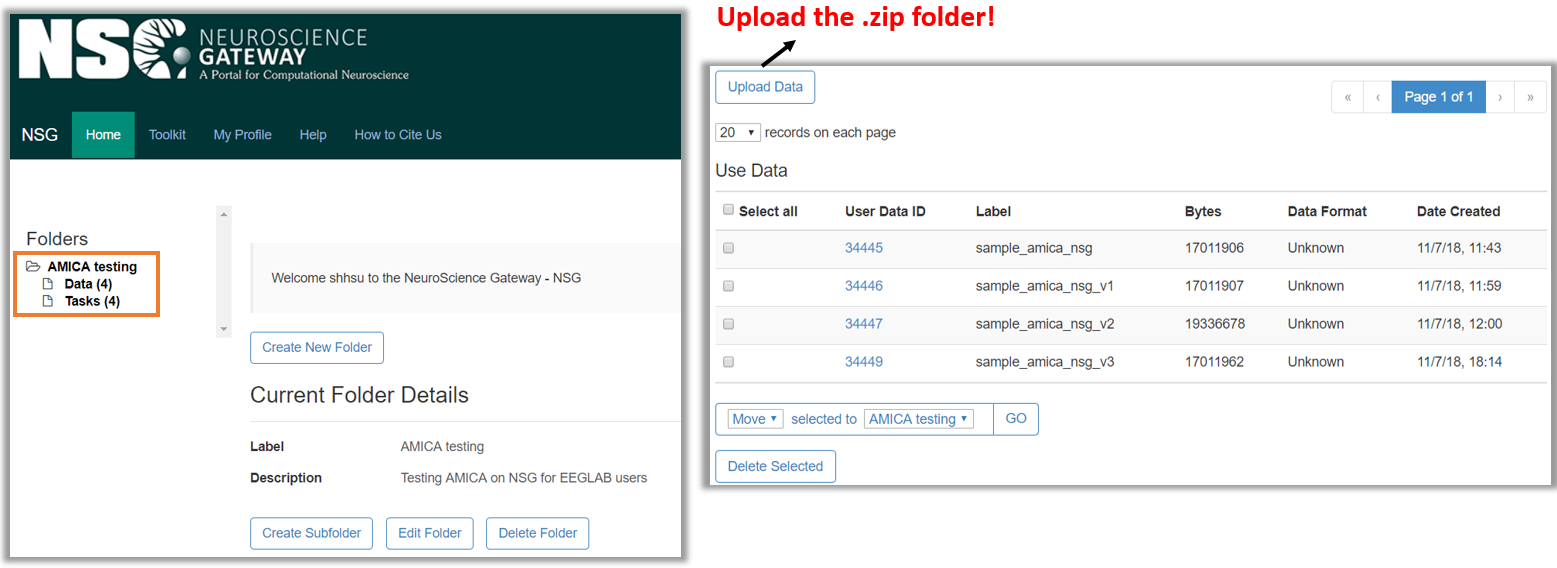
5. Create new task and set input parameters:
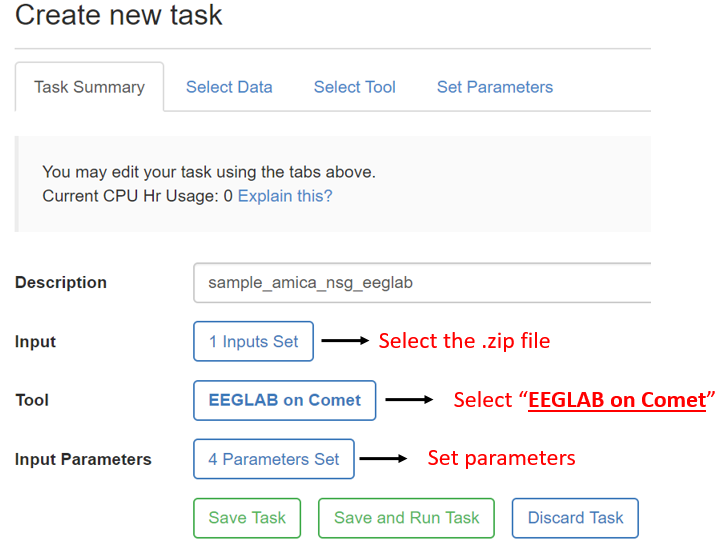
When you click the set parameters button, you will see the tab below:
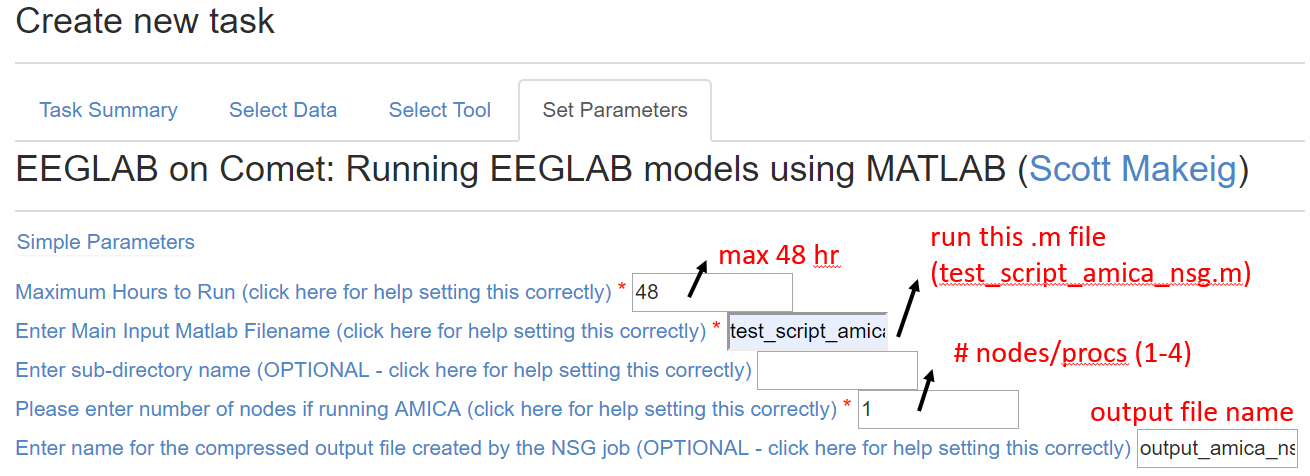
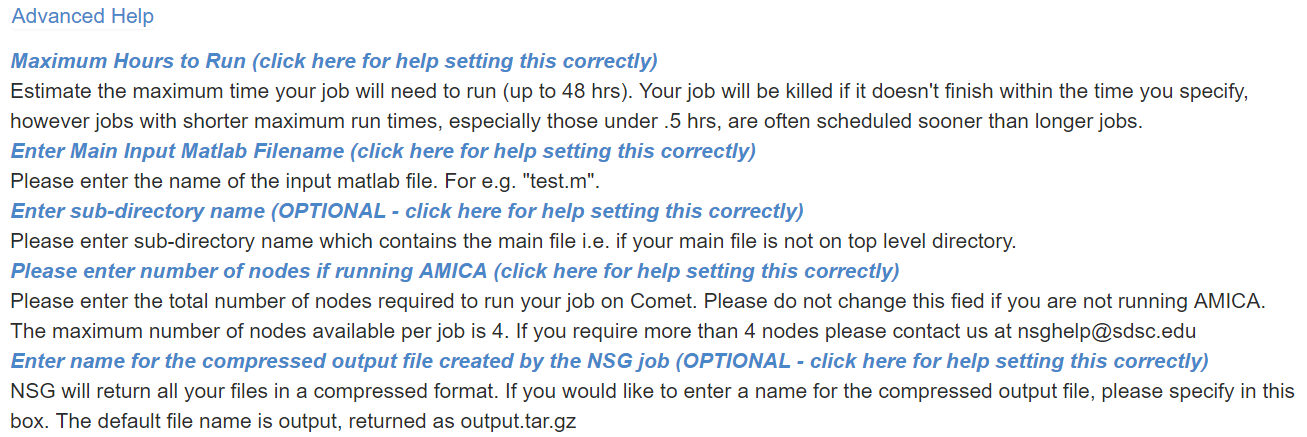
For running AMICA, make sure the number of nodes matches the numprocs in the matlab script because in the NSG back-end different commands are called for single- vs. multiple-nodes tasks. It is also advised to set the max hours to be 48 hours to ensure you get the max available computation time. Click the hyperlink on any of the parameter setting description will lead you to the below advanced help message:
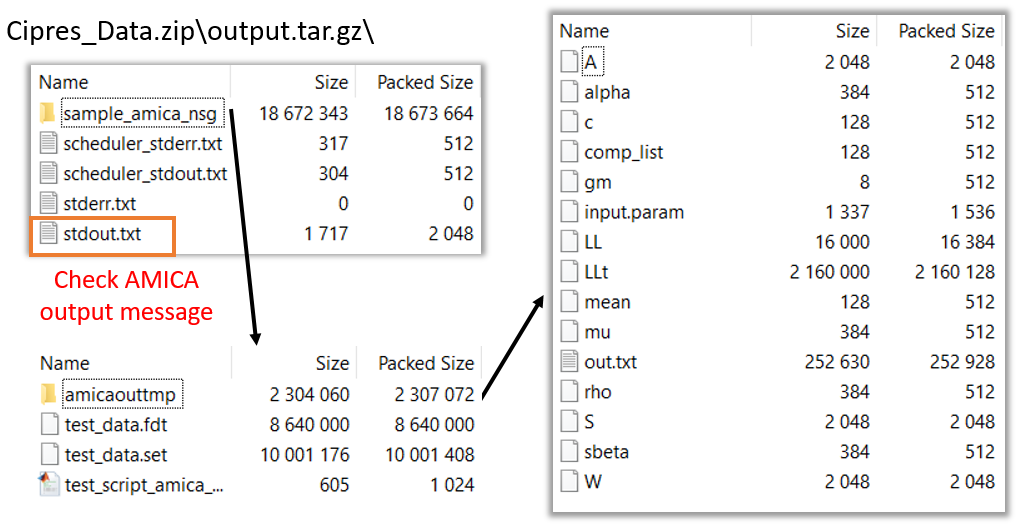
6. Save and run task, view submitted job status, and download the output file: See the EEGLAB on NSG page for detailed instructions.
(Updated 07/15/2019) Now EEGLAB plugin - nsgportal - supports access to NSG. EEGLAB users can now submit and manage jobs to NSG using EEGLAB GUI or command line by installing the nsgportal. For more information and detailed instruction, please visit its Github page.
The AMICA program is capable of scheduling and running on multiple threads. In 2018, the NIH-funded Neuroscience Gateway (NSG) project aims to provide researchers (free) access to computing power on supercomputer clusters to speed up computation-heavy process, including AMICA. With access to 24 cores per node and potentially to multiple nodes on the supercomputer, the runtime of AMICA can be significantly reduced.
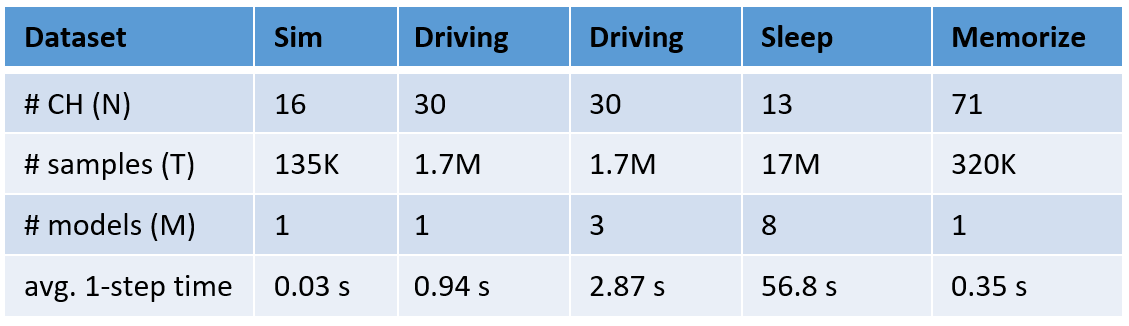
AMICA runtime is proportional to the number of data samples (T) and the total number of parameters to be estimated (M x N^2 + M x N x (4k + 1) + M), where M is the number of models, N is the number of channels, and K is the number of generalized Gaussians for estimating the PDFs.
Analysis for the new Expanse supercomputer is still pending. Below is the runtime analysis of AMICA with multiple threads on Comet, San Diego Supercomputer Center (SDSC):
- Benchmark data: 30-channel EEG, 1.7 million data samples.
- AMICA parameters: default
- Comet computing resources: 24 cores per node. NSG user can access 4 nodes at max for running AMICA.
- Comet CPU spec: Intel Xeon E5-2680v3 processors, 2.5 GHz.
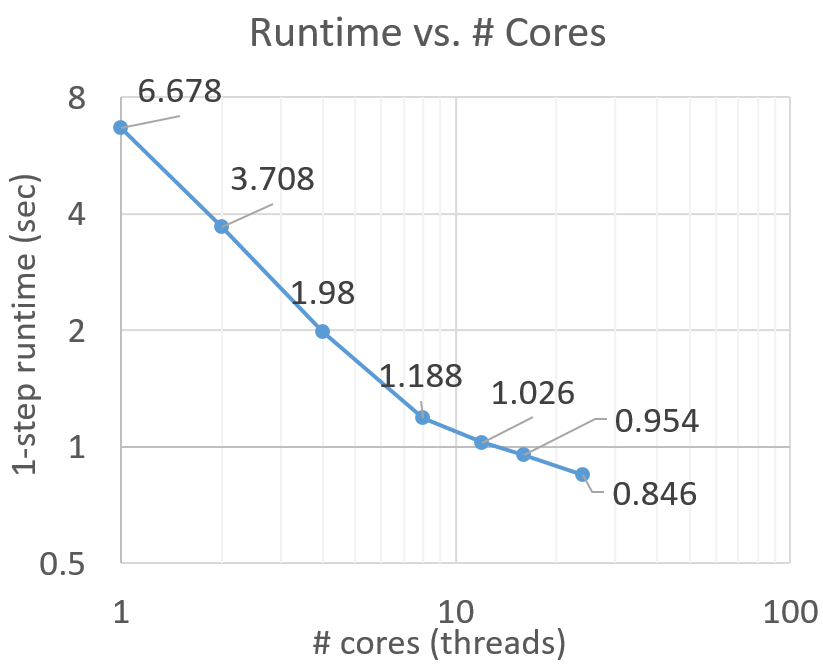

Below is the runtime analysis of AMICA on different datasets with various sizes using 1 node, 24 cores.
EEGLAB users can use postAMICAutility toolbox (download here) for reading and analyzing AMICA output. Please see the AMICA tutorial slides for EEGLAB workshop pp. 22-25 for a quick overview. Detailed description can be found in Hsu, NeuroImage, 2018.