- Example notebooks
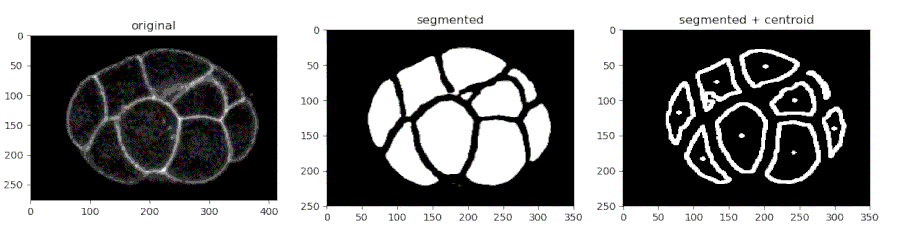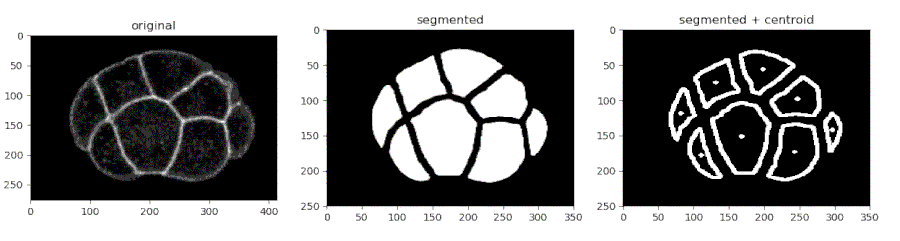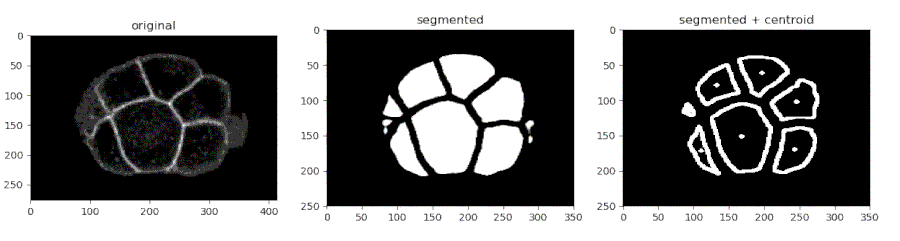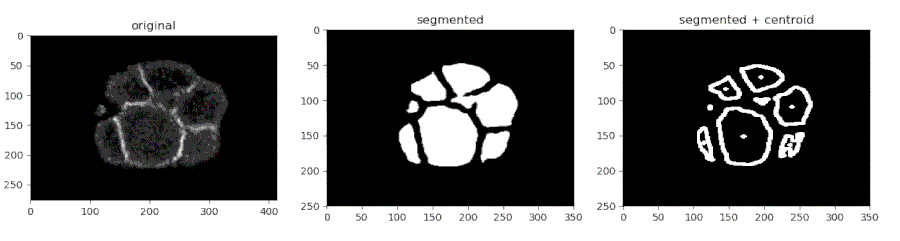
- Segmenting the C. elegans embryo
- Generating synthetic images of embryos with a GAN
- Predicting populations of cells within the C. elegans embryo
- Contributing to DevoLearn
- Links to datasets
- Links to Huggingface spaces
- Contact us
pip install devolearn- Importing the model
from devolearn import cell_membrane_segmentor
segmentor = cell_membrane_segmentor()- Running the model on an image and viewing the prediction
seg_pred = segmentor.predict(image_path = "sample_data/images/seg_sample.jpg")
plt.imshow(seg_pred)
plt.show()- Running the model on a video and saving the predictions into a folder
filenames = segmentor.predict_from_video(video_path = "sample_data/videos/seg_sample.mov", centroid_mode = False, save_folder = "preds")- Finding the centroids of the segmented features
seg_pred, centroids = segmentor.predict(image_path = "sample_data/images/seg_sample.jpg", centroid_mode = True)
plt.imshow(seg_pred)
plt.show()- Saving the centroids from each frame into a CSV
df = segmentor.predict_from_video(video_path = "sample_data/videos/seg_sample.mov", centroid_mode = True, save_folder = "preds")
df.to_csv("centroids.csv")- Importing the model
from devolearn import cell_nucleus_segmentor
segmentor = cell_nucleus_segmentor()- Running the model on an image and viewing the prediction
seg_pred = segmentor.predict(image_path = "sample_data/images/nucleus_seg_sample.jpg")
plt.imshow(seg_pred)
plt.show()- Importing the model
from devolearn import Generator, embryo_generator_model
generator = embryo_generator_model()- Generating a picture and viewing it with matplotlib
gen_image = generator.generate()
plt.imshow(gen_image)
plt.show()- Generating n images and saving them into
foldernamewith a custom size
generator.generate_n_images(n = 5, foldername= "generated_images", image_size= (700,500))- Importing the population model for inferences
from devolearn import lineage_population_model- Loading a model instance to be used to estimate lineage populations of embryos from videos/photos.
model = lineage_population_model(device = "cpu")- Making a prediction from an image
print(model.predict(image_path = "sample_data/images/embryo_sample.png"))- Making predictions from a video and saving the predictions into a CSV file
results = model.predict_from_video(video_path = "sample_data/videos/embryo_timelapse.mov", save_csv = True, csv_name = "video_preds.csv", ignore_first_n_frames= 10, ignore_last_n_frames= 10, postprocess = False)- Plotting the model's predictions from a video
plot = model.create_population_plot_from_video(video_path = "sample_data/videos/embryo_timelapse.mov", save_plot= True, plot_name= "plot.png", ignore_last_n_frames= 0, postprocess = False)
plot.show()| Model | Data source |
|---|---|
| Segmenting the cell membrane in C. elegans embryo | 3DMMS: robust 3D Membrane Morphological Segmentation of C. elegans embryo |
| Segmenting the nucleus in C. elegans embryo | C. elegans Cell-Tracking-Challenge dataset |
| Cell lineage population prediction + embryo GAN | EPIC dataset |
| Model | Huggingface |
|---|---|
| Segmenting the cell membrane in C. elegans embryo | Cell Membrane segmentor |
| Segmenting the nucleus in C. elegans embryo | C. elegans Nucleus segmentor |
| Cell lineage population prediction | Lineage population |
Feel free to join our Slack workspace!





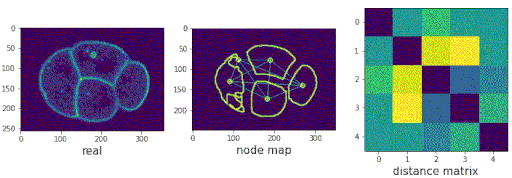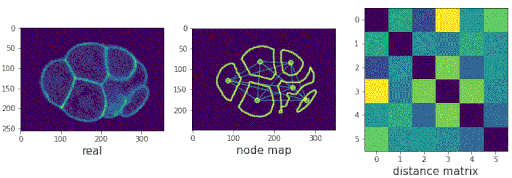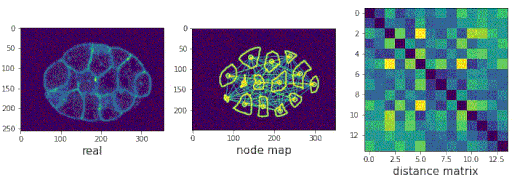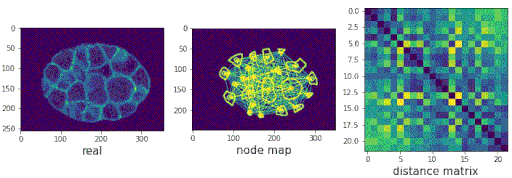

/images/generated_embryos_3.gif)
/images/resnet_preds_with_input.gif)